|
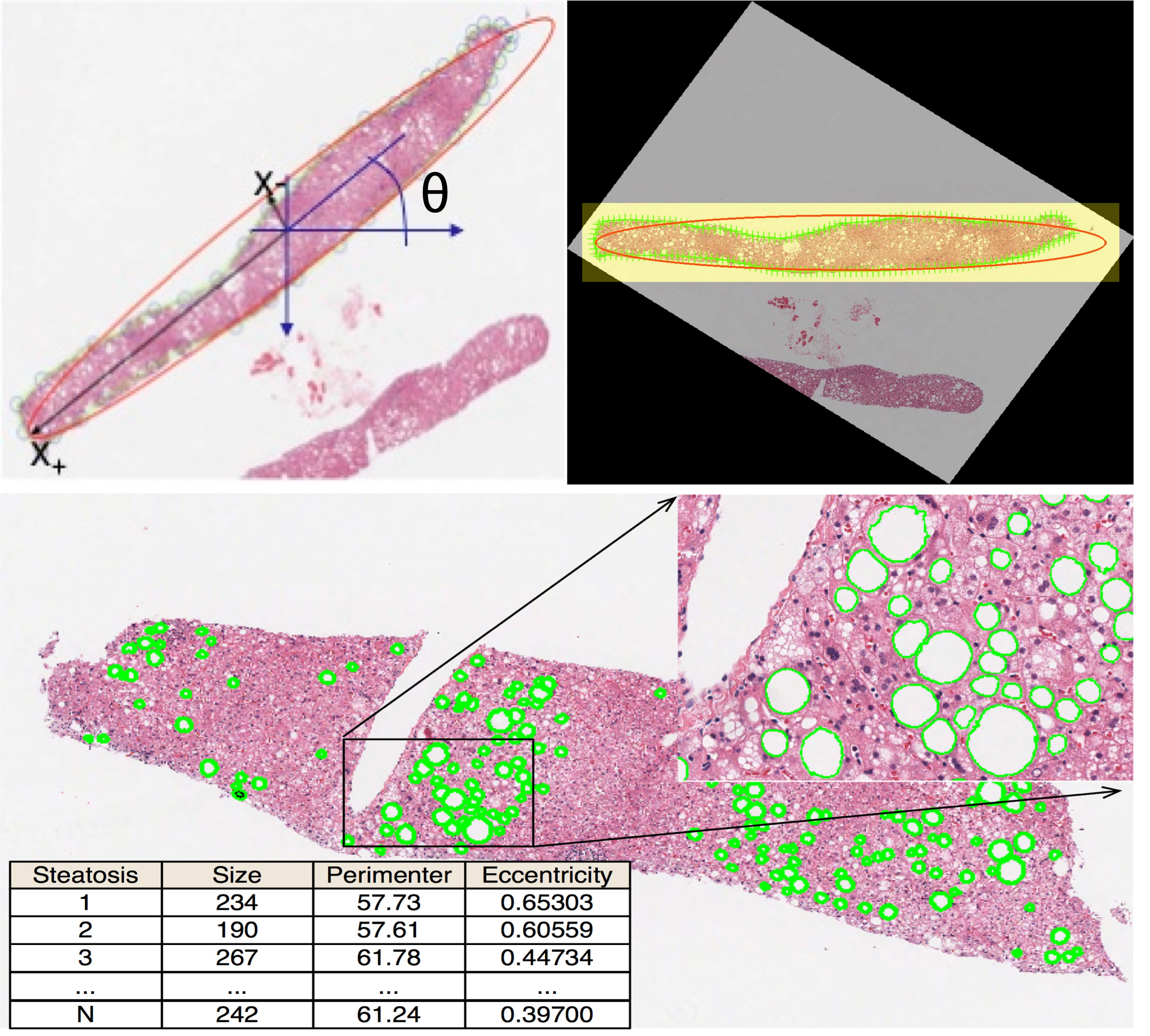
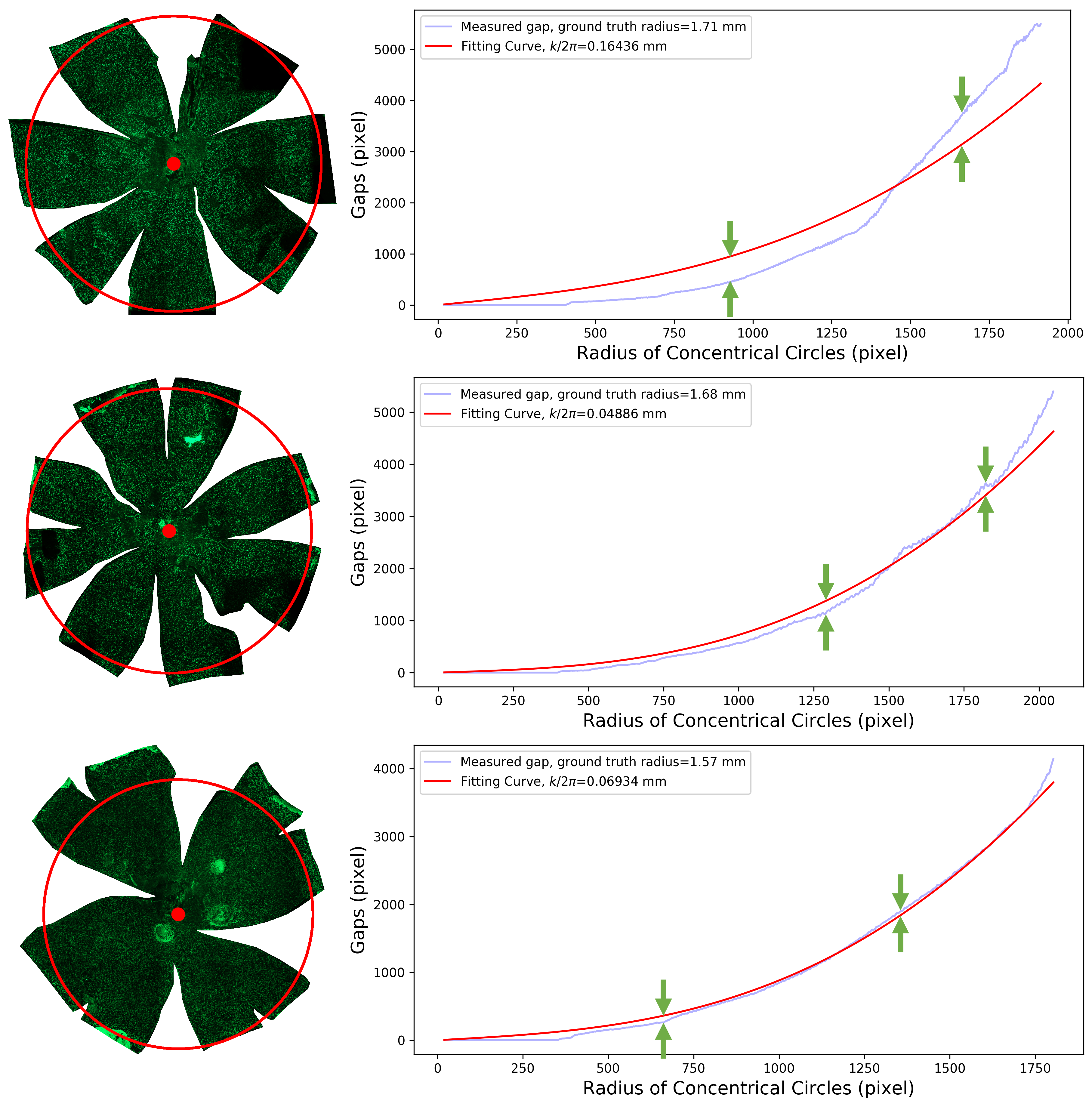
-- Retinal Pigment Epithelial (RPE) cells serve as a supporter for the metabolism and visual function of photoreceptors and a barrier for photoreceptor protection. Morphology dynamics, spatial organization, distribution density, and growth patterns of RPE cells are important for further research on these RPE main functions. To enable such investigations within the authentic eyeball structure, we present a new method to estimate the three-dimension (3D) eyeball sphere from two-dimension (2D) tissue flat-mount microscopy images. An error-correction term is formulated to compensate for the reconstruction error due to tissue distortions. We evaluate the effect of the tissue-distortion error by excluding partial data points from the low- and high-latitude zones. The error-correction parameter is learned automatically using a set of samples with the ground truth eyeball diameters gauged by noncontact LED micrometry at sub-micron accuracy and precision. Our analysis demonstrates that the error-correction term in the reconstruction model is a valid way to model tissue distortions in the tissue flat-mount preparation steps. We have developed a new method to enable RPE morphometry analysis with respect to locations on an eyeball sphere, an important step to further enhance RPE research and eye disease diagnosis. 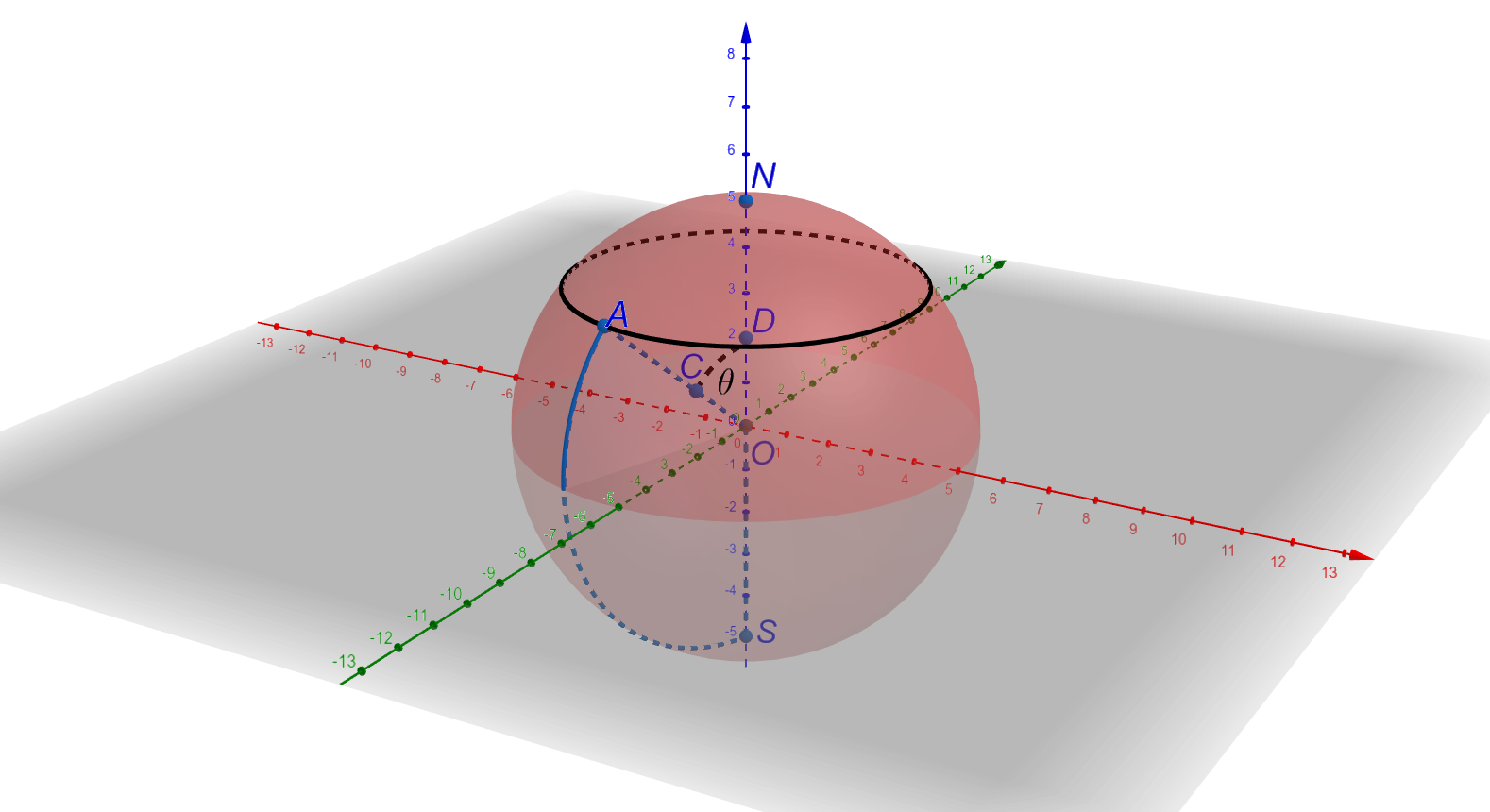
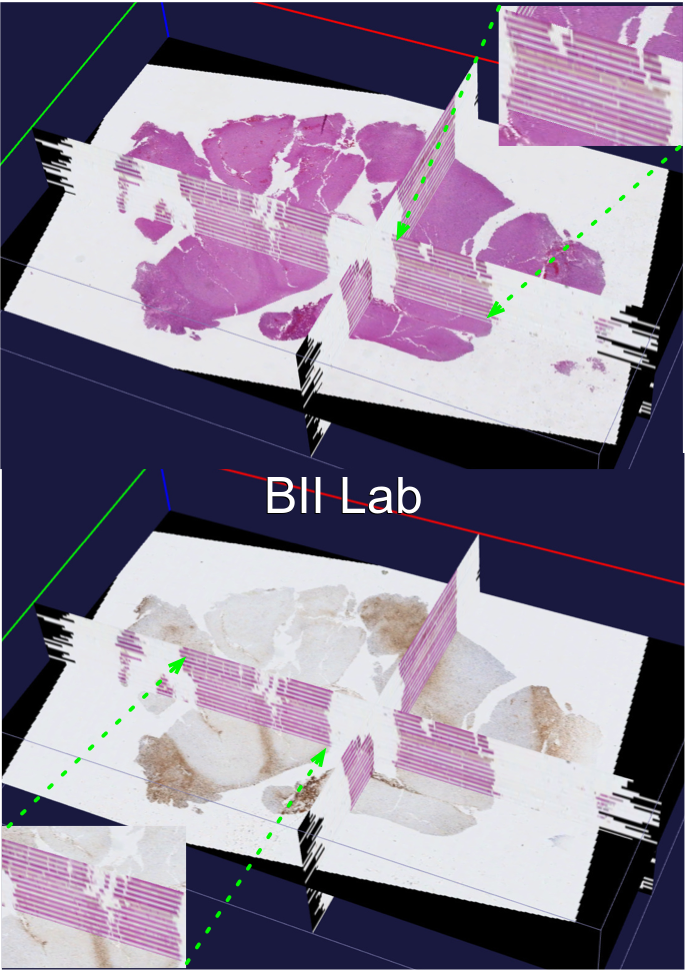
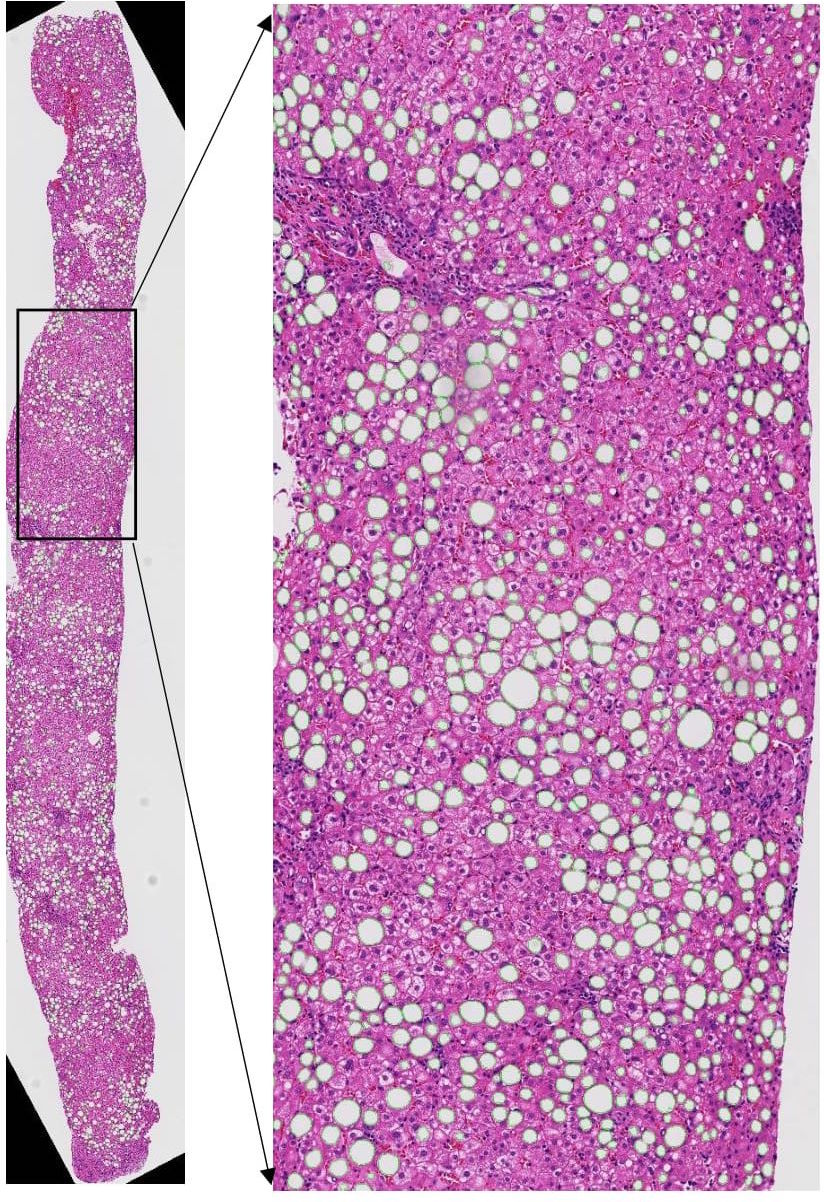
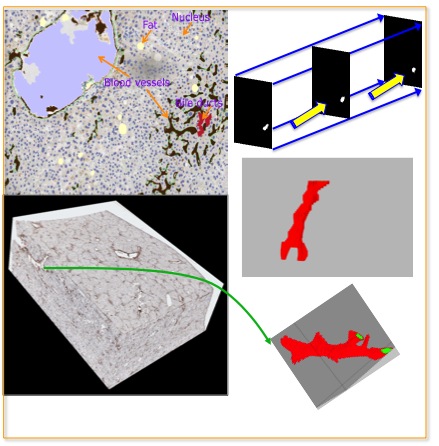
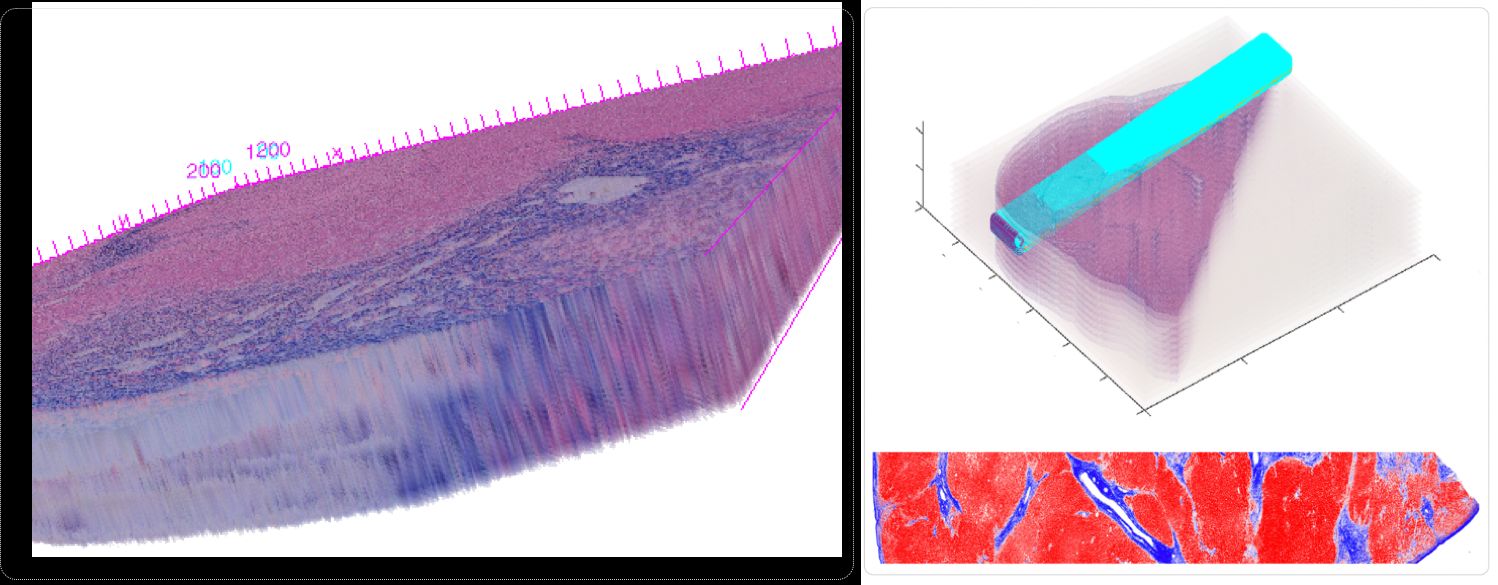
-- In spite of serving as the "gold-standard" for diagnosis of hepatic fibrosis and cirrhosis, the liver needle biopsy is known to include inaccurate diagnostic information, primarily due to the inexorable sampling bias. However, there is lack of method to gauge such sampling bias as it is infeasible to take a large number of needle biopsies from the same patient. To develop a computational analysis pipeline to enable needle biopsy sampling bias investigations, we construct a 3D virtual liver tissue volume by spatially registered high resolution Whole Slide Images (WSIs) of serial liver tissue sections with a novel dynamic registration method. With spatially aligned serial liver WSIs, we further develop a Virtual Needle Biopsy Sampling function (VNBS) that mimics the needle biopsy sampling process with a physical needle in current clinical practice. Such a virtual sampling function is applied to the reconstructed digital liver image volume for multiple times, each at a distinct tissue location and angle. Additionally, we develop an analysis pipeline to quantitate Collagen Proportionate Area (CPA) in all resulting needle biopsies. All sampled biopsies are manually graded by the Scheuer and Ishak staging methods. With such domain annotations and machine-based collagen fiber quantification results, we investigate the inter- and intra-biopsy variability by the pathology stage score and CPA with a set of 3D virtual needle biopsies and their internal 2D cross sections. Our experimental results suggest the developed computational pipeline can recover the substantial presence of needle biopsy sampling bias in both pathology stage scores and liver CPAs.The developed virtual liver needle biopsy sampling method provides a new avenue for investigating needle biopsy sampling bias with 3D virtual tissue volumes. Although the developed working pipeline specifically investigates the sampling bias in liver needle biopsy in this study, the presented analysis methods can be tailored to a large scope of other tissue based disease diagnoses where the needle biopsy sampling bias substantially affects the diagnostic results. 
-- In the era of precision medicine, human tumor atlas oriented studies have been significantly facilitated by high-resolution, multi-modal tissue based microscopic pathology image analytics. To better support such tissue-based investigations, we develop Digital Pathology Laboratory (DPLab), a publicly available web-based platform, to assist biomedical research groups, non-technical end users, and clinicians for pathology Whole-Slide Image (WSI) visualization, annotation, analysis, and sharing via web browsers. A major advance of this work is the easy-to-follow methods to reconstruct three-dimension (3D) tissue image volumes by registering two-dimension (2D) whole-slide pathology images of serial tissue sections stained by hematoxylin and eosin (H&E), and immunohistochemistry (IHC). The integration of these serial slides stained by different methods provides cellular phenotype and pathophysiologic states in the context of a 3D tissue micro-environment. DPLab is hosted on a publicly accessible server and connected to a backend computational cluster for intensive image analysis computations, with results visualized, downloaded, and shared via a web interface. Equipped with an analysis toolbox of numerous image processing algorithms, DPLab supports continued integration of community-contributed algorithms and presents an effective solution to improve the accessibility and dissemination of image analysis algorithms by research communities. It represents the first step in making next generation tissue investigation tools widely available to the research community, enabling and facilitating discovery of clinically relevant disease mechanisms in a digital 3D tissue space. 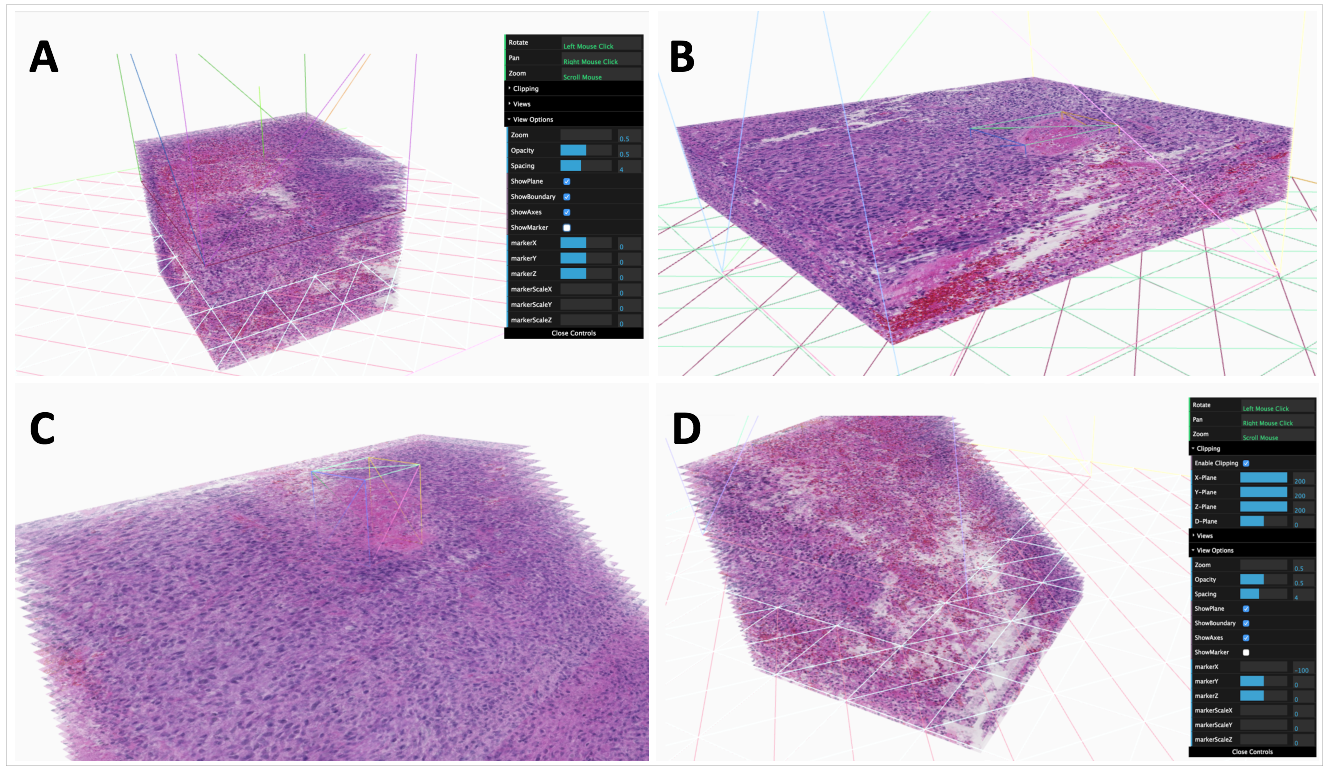
|
Hongxiao Li, Hanyi Yu, Yong-Kyu Kim, Fusheng Wang, George Teodoro, Yi Jiang, John Nickerson, Jun Kong, Computational Model-Based Estimation of Mouse Eyeball Structure From Two-Dimensional Flatmount Microscopy Images, Translational Vision Science and Technology,10(4):25, 2021.
Qiang Li, Fusheng Wang, Yaobing Chen, Hao Chen, Shengdi Wu, Alton Farris, Yi Jiang, Jun Kong, Virtual Liver Needle Biopsy from Reconstructed Three-Dimensional Histopathological Images: Quantification of Sampling Error, Computers in Biology and Medicine, Volume 147, 105764, 2022.
Alice Shen, Fusheng Wang, Saptarshi Paul, Divya Bhuvanapalli, Jacob Alayof, Alton B. Farris, George Teodoro, Daniel J. Brat, Jun Kong, An integrative web-based software tool for multi-dimensional pathology whole-slide image analytics, Physics in Medicine and Biology, Accepted, 2022
|
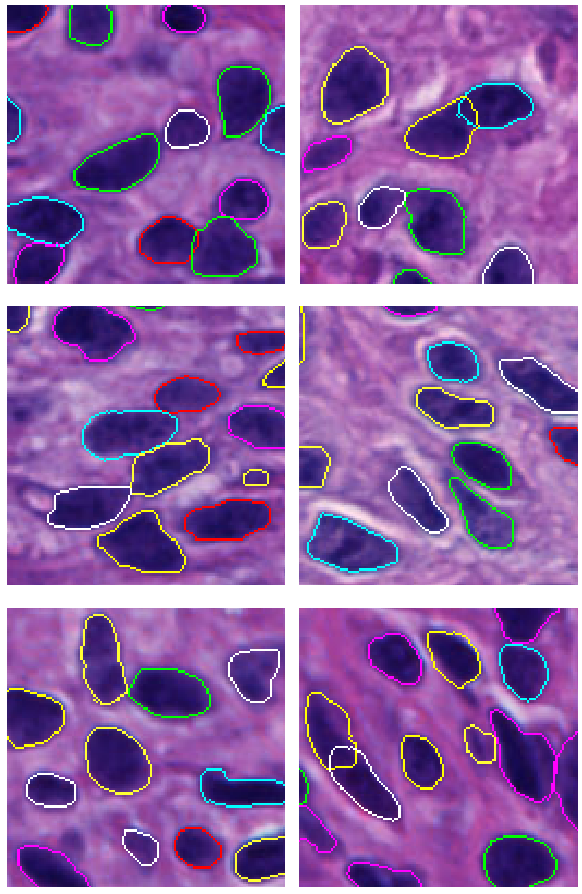
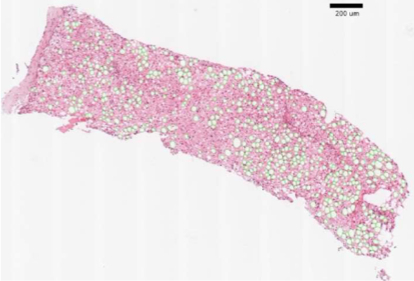
-- Retinal pigment epithelium (RPE) aging is an important cause of vision loss. As RPE aging is accompanied by changes in cell morphological features, an accurate segmentation of RPE cells is a prerequisite to such morphology analyses. Due the overwhelmingly large cell number, manual annotations of RPE cell borders are time-consuming. Computer based methods do not work well on cells with weak or missing borders in the impaired RPE sheet regions. To address such a challenge, we develop a semi-supervised deep learning approach, namely MultiHeadGAN, to segment low contrast cells from impaired regions in RPE flatmount images. The developed deep learning model has a multi-head structure that allows model training with only a small scale of human annotated data. To strengthen model learning effect, we further train our model with RPE cells without ground truth cell borders by generative adversarial networks. Additionally, we develop a new shape loss to guide the network to produce closed cell borders in the segmentation results. Compared with other state-of-the-art deep learning approaches, our method demonstrates superior qualitative and quantitative performance.Suggested by our extensive experiments, our developed deep learning method can accurately segment cells from RPE flatmount microscopy images and is promising to support large scale cell morphological analyses for RPE aging investigations. 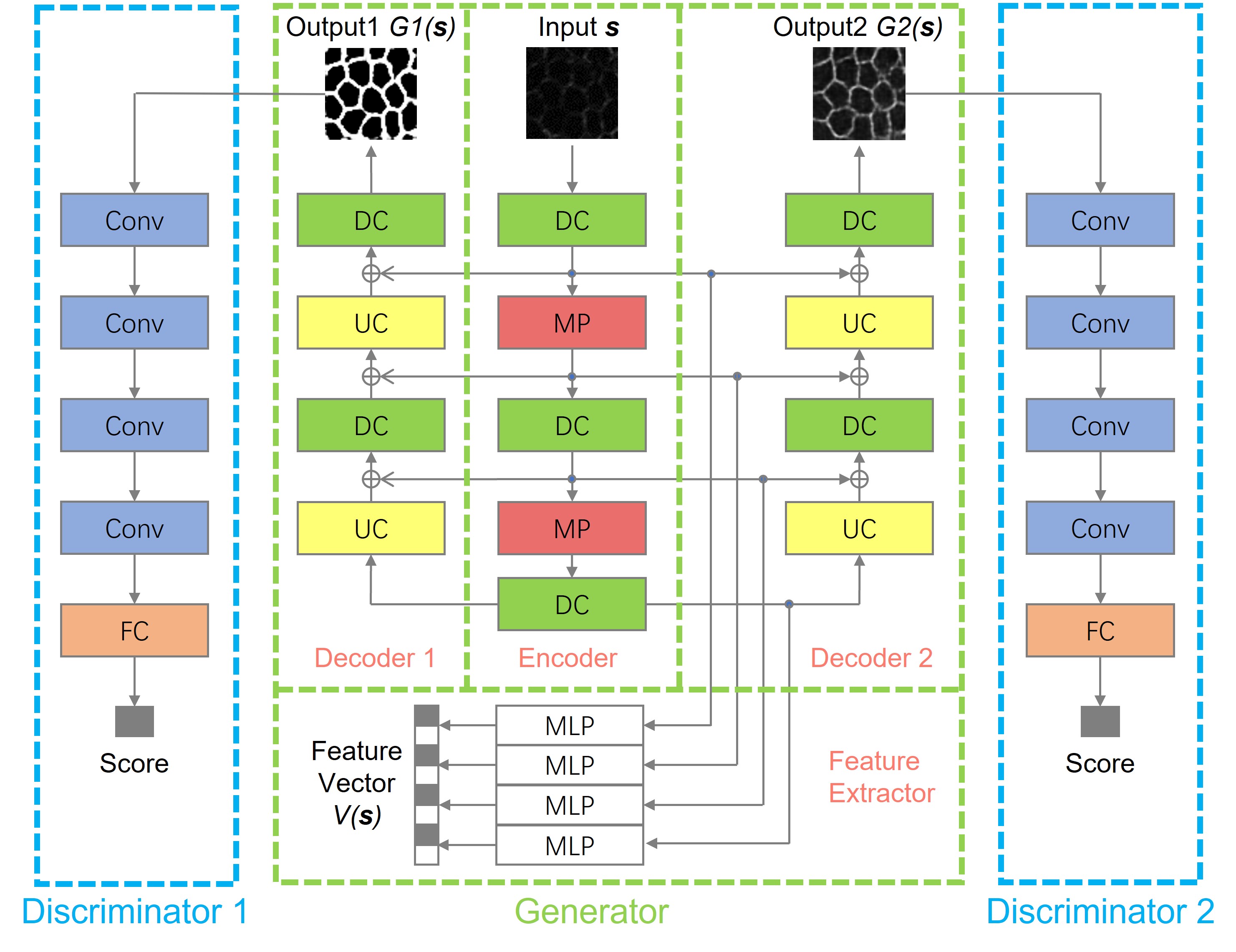
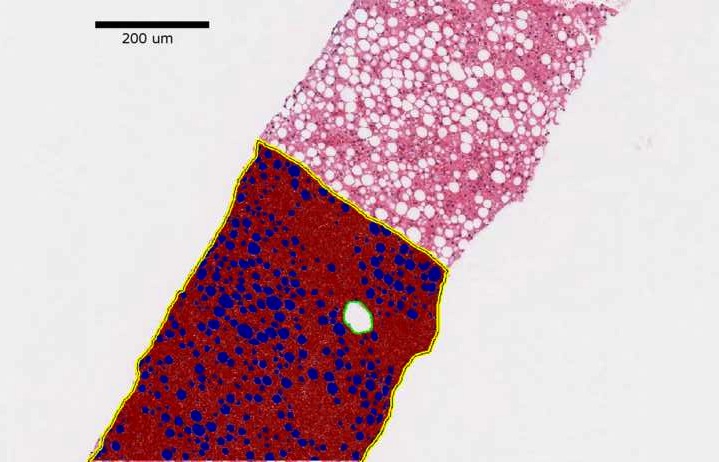
-- Liver fibrosis staging is clinically important for liver disease progression prediction. As the portal tract fibrotic quantity and size in a liver biopsy correlate with the fibrosis stage, an accurate analysis of portal tract regions is clinically critical. Manual annotations of portal tract regions, however, are time-consuming and subject to large inter- and intra-observer variability. To address such a challenge, we develop a Multi-up-sampling and Spatial Attention guided UNet model (MUSA-UNet) to segment liver portal tract regions in whole-slide images of liver tissue slides. To enhance the segmentation performance, we propose to use depth-wise separable convolution, the spatial attention mechanism, the residual connection, and multiple up-sampling paths in the developed model. Compared with other state-of-the-art deep learning methods, our model demonstrates both superior qualitative and quantitative performance. The clinical Scheuer fibrosis stage presents a strong correlation with the resulting average portal tract fibrotic area and portal tract percentage computed from the MUSA-UNet segmentation results. In conclusion, our developed deep learning model MUSA-UNet can accurately segment portal tract regions from whole-slide images of liver tissue biopsies and present a promising potential to assist liver disease diagnosis in a computational manner. 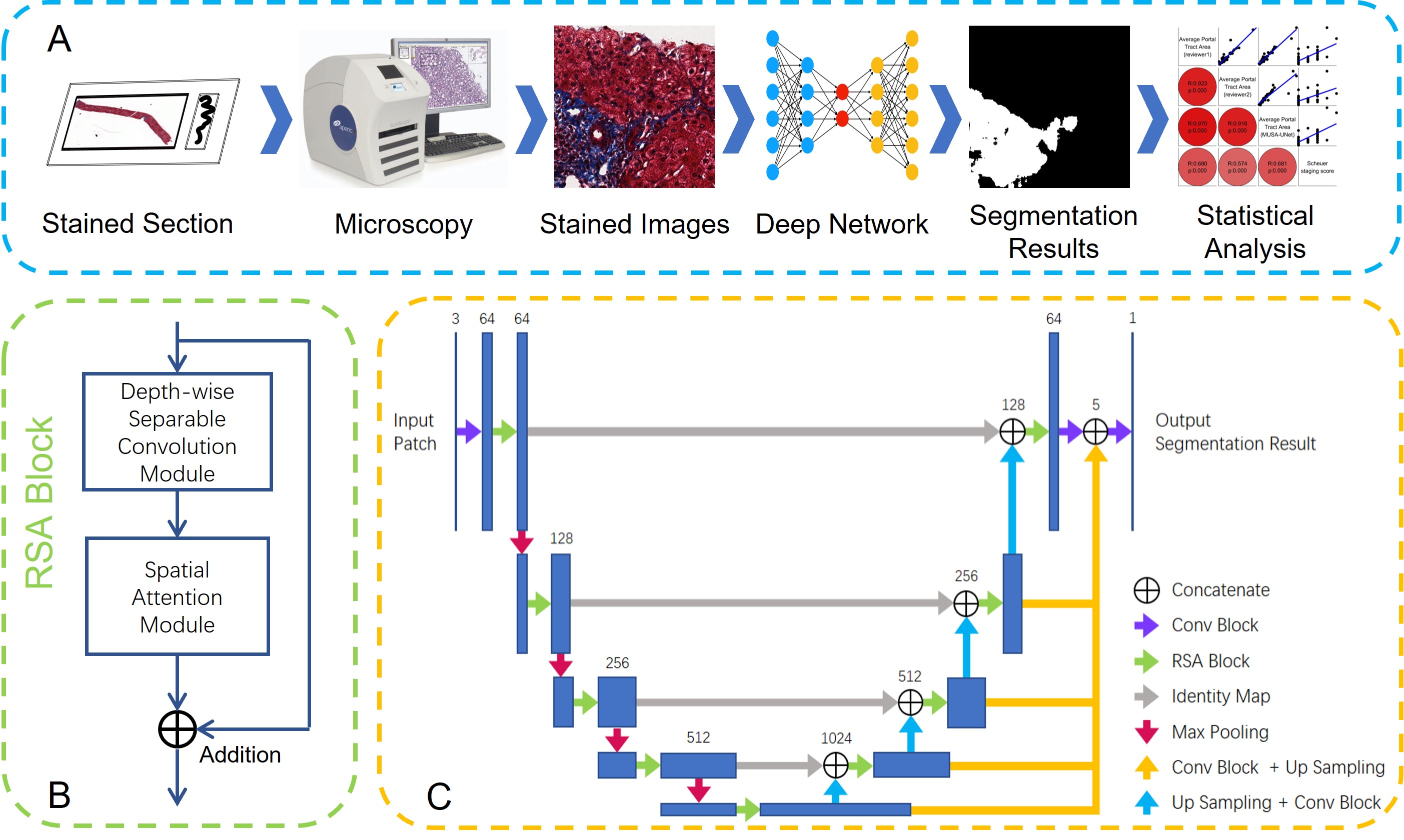
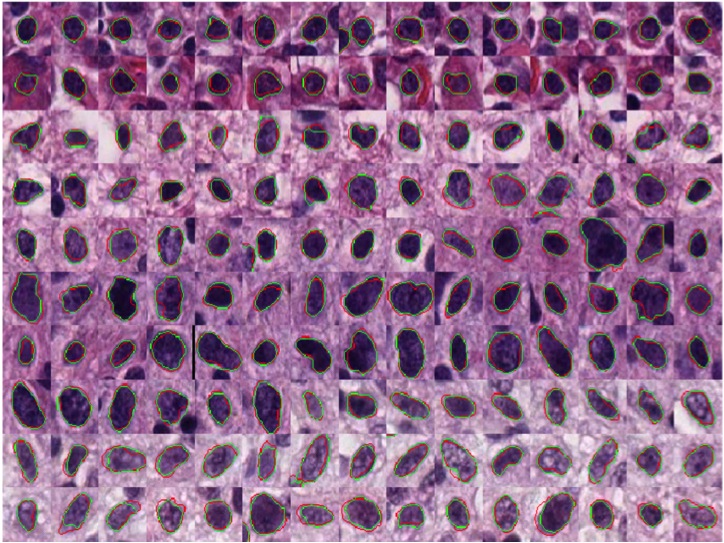
-- Pathology image analysis is a crucial step to support accurate cancer mechanism research, disease grading, diagnosis, and treatment planning. Although there is a high demand for precise nuclei morphology analyses, nuclei segmentation, the prerequisite step, mostly is completed manually by pathologists in practice. While the newly emerging deep learning algorithms can help automate the process, the lack of sufficient pathology training images with well-annotated ground truth in most tissue-based biomedical research inevitably limits the performance of deep learning systems. In this study, we propose to address this problem by a convolutional neural network with foveal blur, a blurring algorithm mimicking human vision, that enriches datasets with multiple local nuclei regions of interest derived from large pathology images for enhanced nuclei segmentation performance. As the convolutional neural network is not able to explicitly learn the nuclei shape, we propose a human knowledge boosted deep learning system by inclusion to the convolutional neural network new loss function terms capturing shape prior knowledge and imposing smoothness constraints. The high segmentation accuracy and competitive processing speed of our proposed method from three independent datasets suggest its promising potential for automating histopathology nuclei segmentation in biomedical research and clinical settings where efficient high-quality nuclei segmentation are demanded. 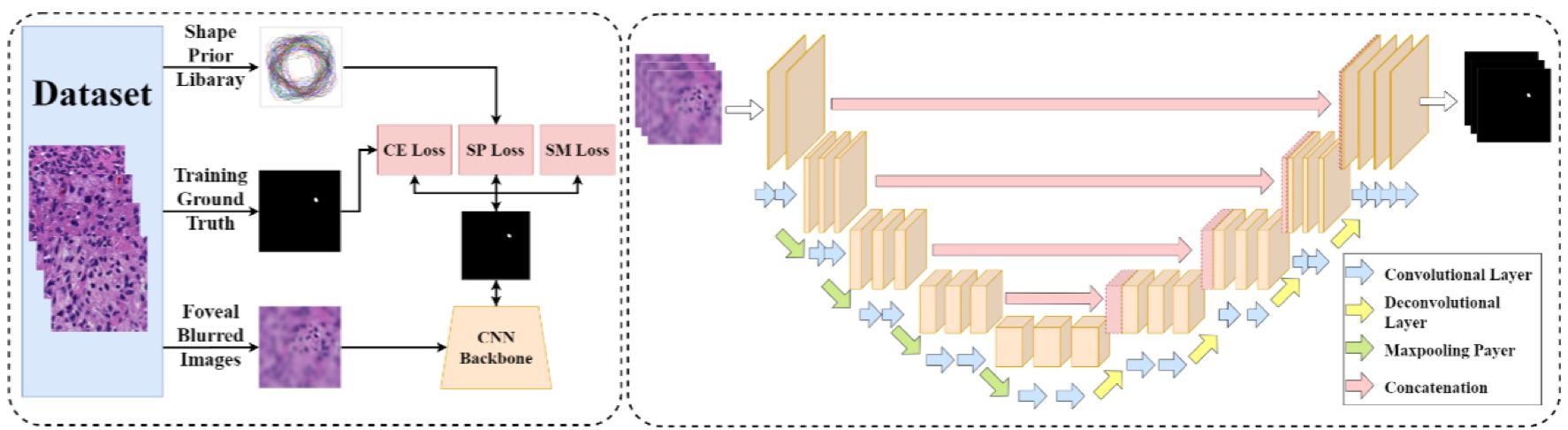
-- In this study, we present a multi-resolution deep learning model HistoCAE for viable tumor segmentation in whole-slide liver histopathology images. We propose convolutional autoencoder (CAE) based framework with a customized reconstruction loss function for image reconstruction, followed by a classification module to classify each image patch as tumor vs non-tumor. The resulting patch-based prediction results are spatially combined to generate the final segmentation result for each WSI. Our proposed model presents superior performance to other benchmark models with extensive experiments, suggesting its efficacy for viable tumor area segmentation with liver whole-slide images. 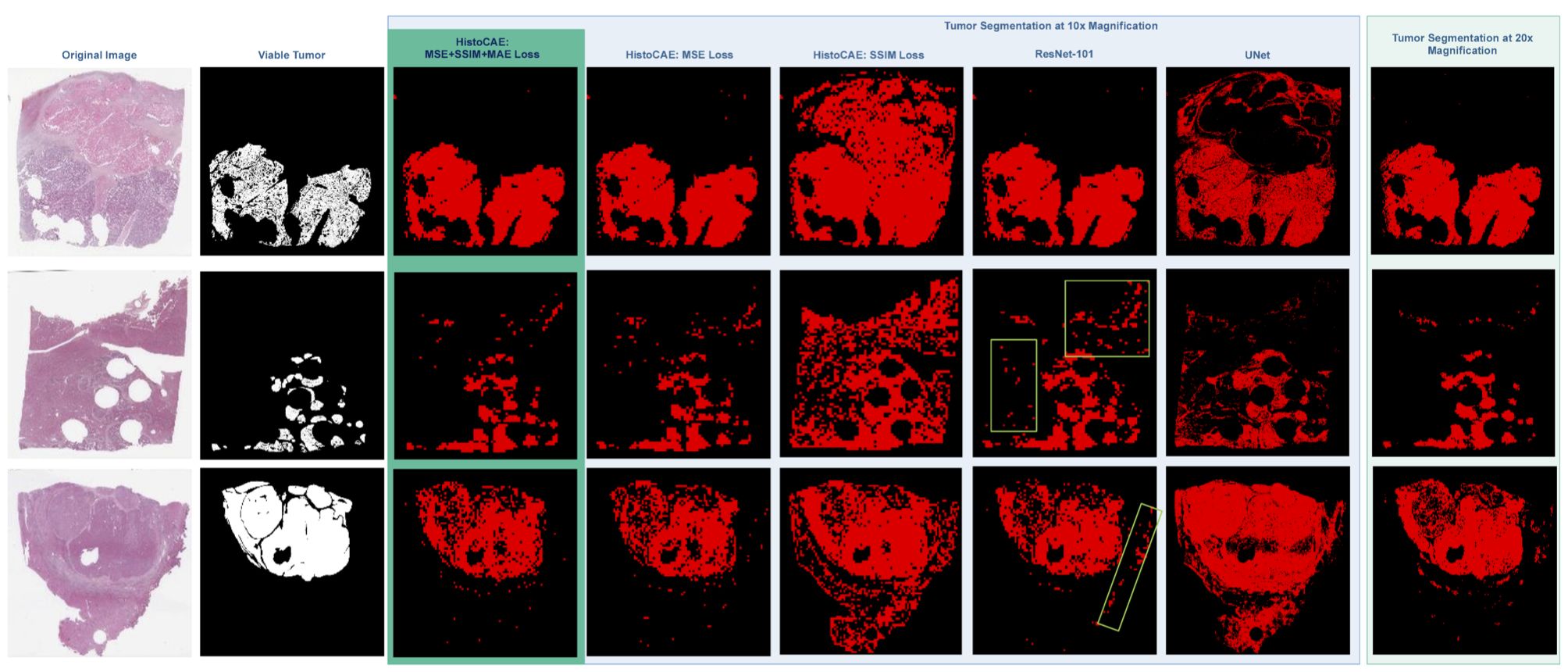
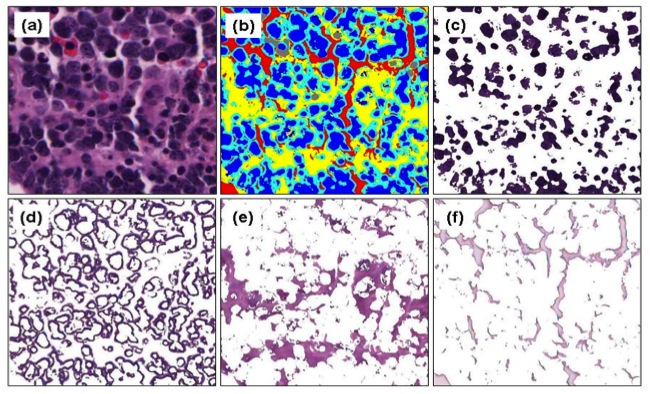
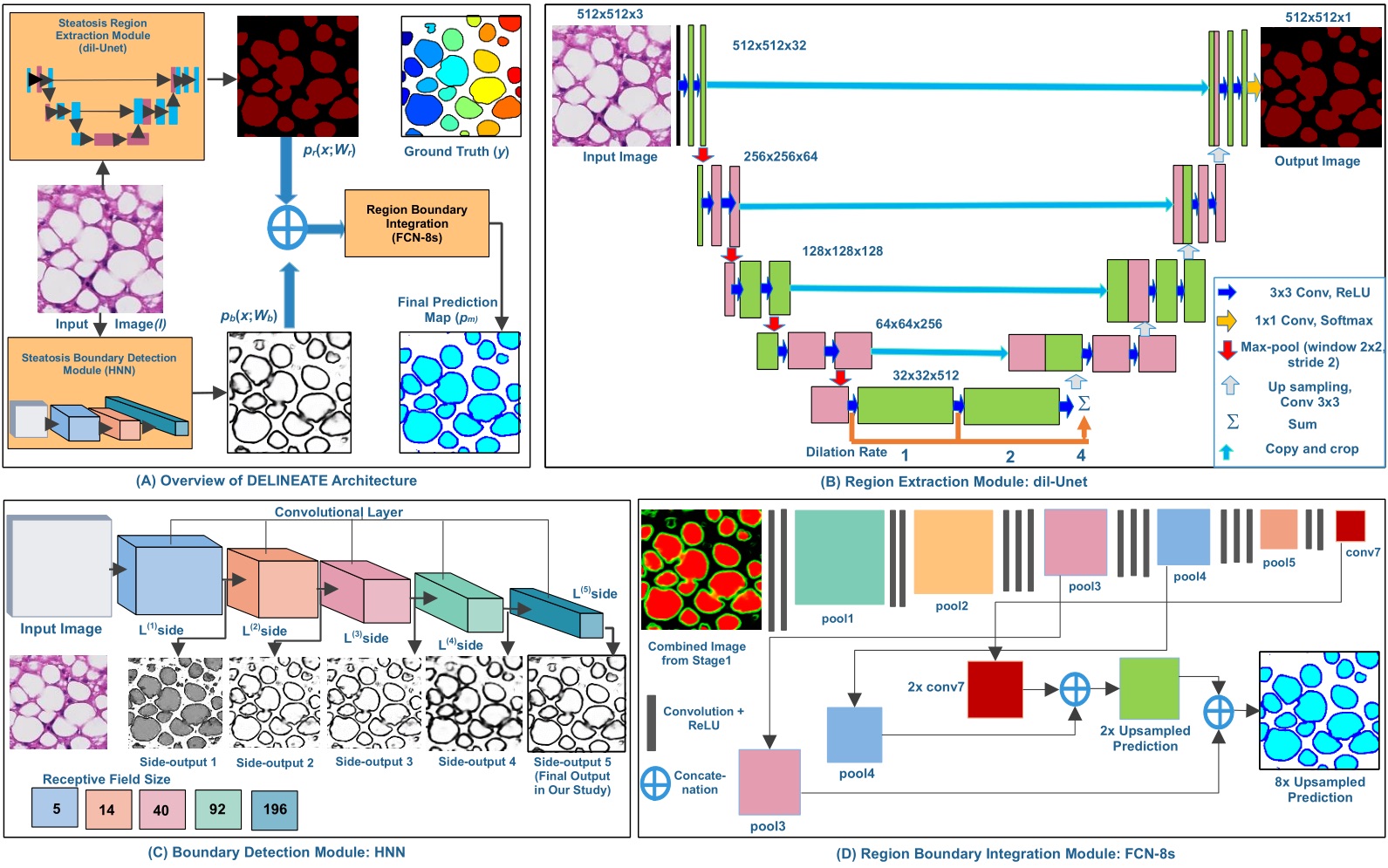
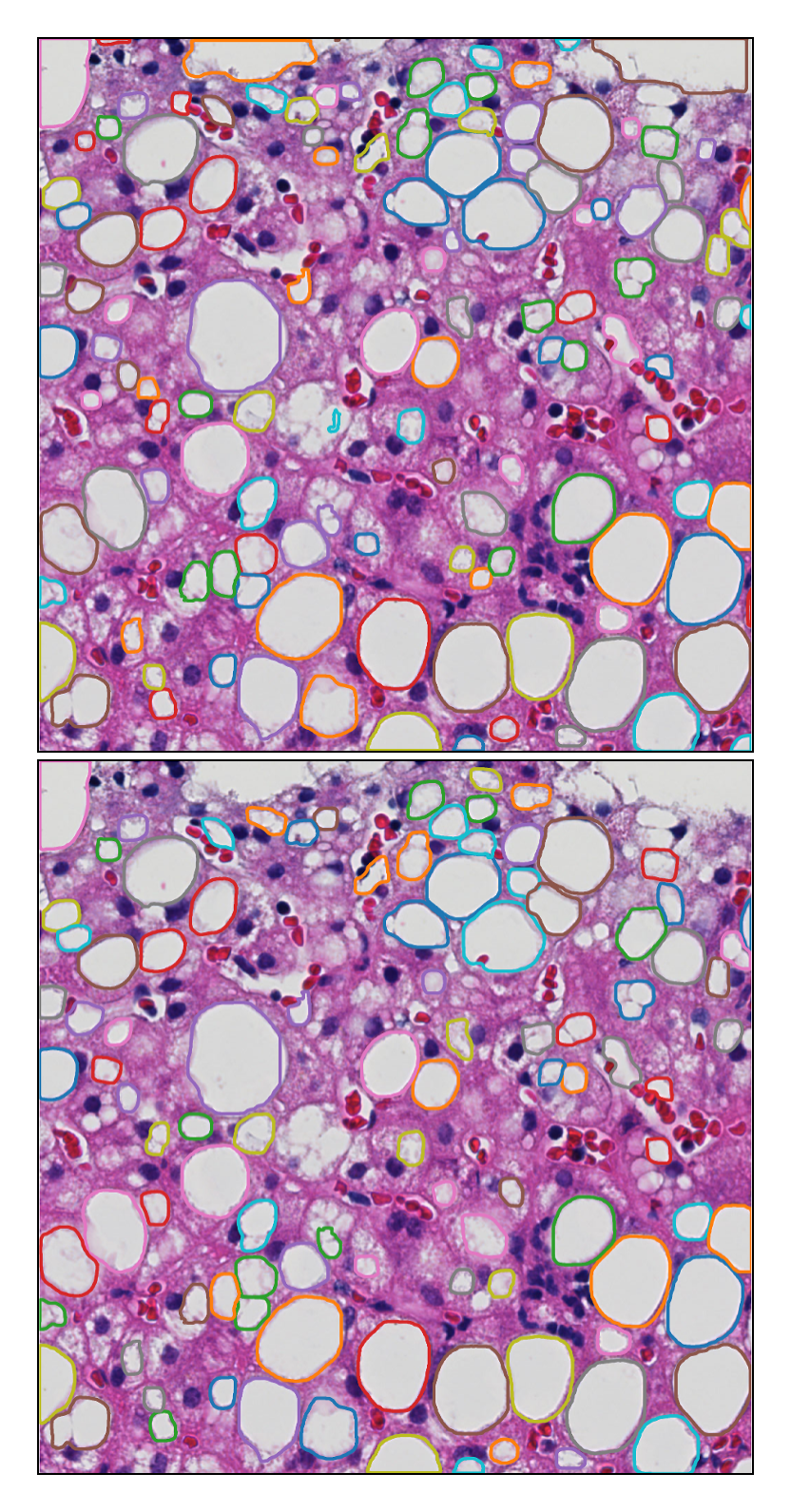
-- Liver steatosis is known as the abnormal accumulation of lipids within cells. An accurate quantification of steatosis area within the liver histopathological microscopy images plays an important role in liver disease diagnosis and trans- plantation assessment. Such a quantification analysis often requires a precise steatosis segmentation that is challenging due to abundant presence of highly overlapped steatosis droplets. In this paper, a deep learning model Mask-RCNN is used to segment the steatosis droplets in clumps. Extended from Faster R-CNN, Mask-RCNN can predict object masks in addition to bounding box detection. With transfer learning, the resulting model is promising to support liver disease diagnosis and allograft rejection prediction in future clinical practice. 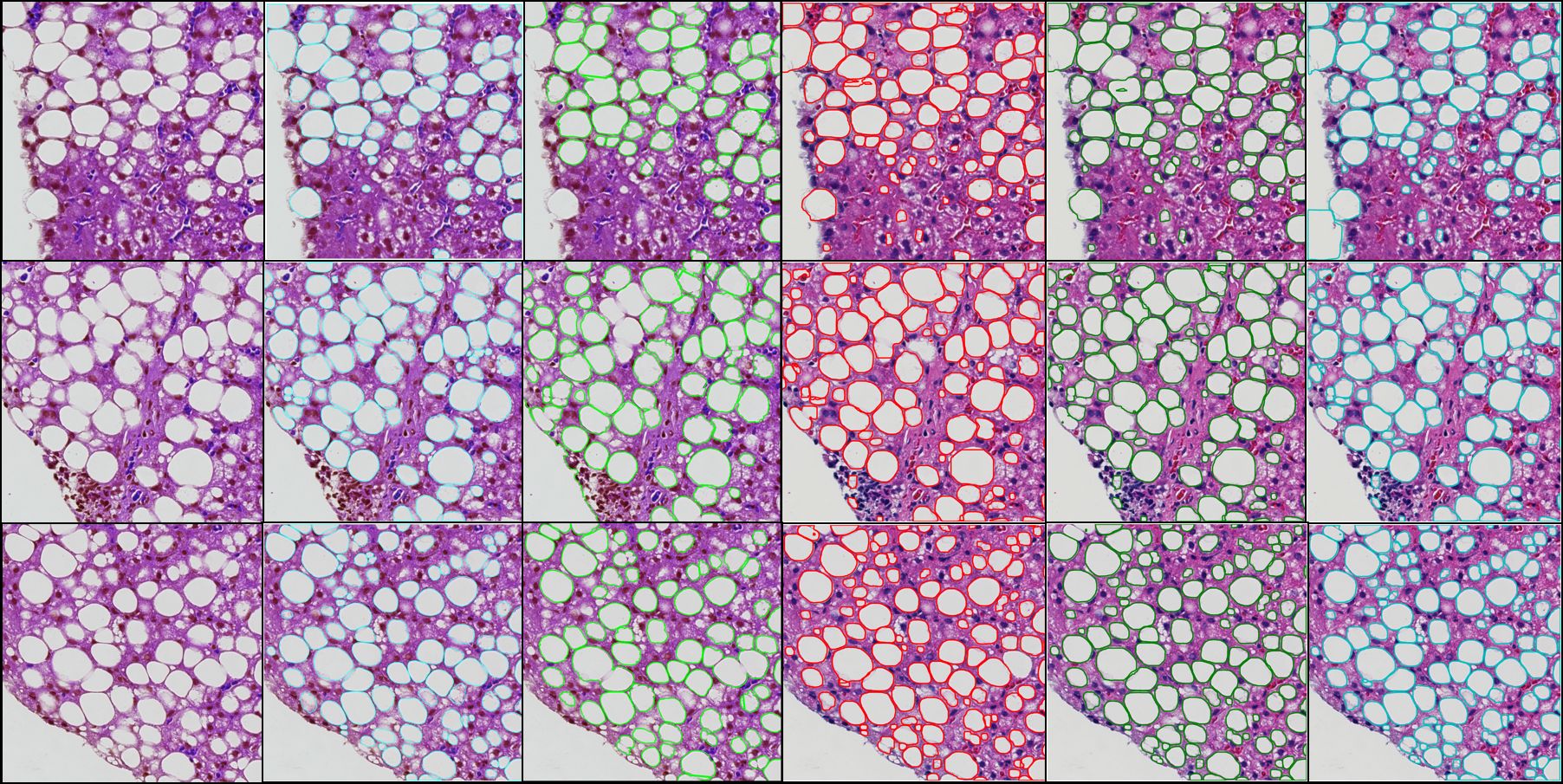
|
Hanyi Yu, Fusheng Wang, George Teodoro, John Nickerson, Jun Kong, MultiHeadGAN: A deep learning method for low contrast retinal pigment epithelium cell segmentation with fluorescent flatmount microscopy images, Computers in Biology and Medicine, Volume 146, 105596, 2022.
Hanyi Yu, Nima Sharifai, Kun Jiang, Fusheng Wang, George Teodoro, Alton B. Farris, Jun Kong, Artifi- cial Intelligence based Liver Portal Tract Region Identification and Quantification with Transplant Biopsy Whole-Slide Images,” Computers in Biology and Medicine, Vol 150, 106089, 2022.
Hongyi Duanmu, Fusheng Wang, George Teodoro, Jun Kong, Foveal Blur-Boosted Segmentation of Nuclei in Histopathology Images with Shape Prior Knowledge and Constraints, Bioinformatics, Volume 37, Issue 21, pp.3905-3913, 2021.
Mousumi Roy, Jun Kong, Satyananda Kashyap, Vito Paolo Pastore, Fusheng Wang, Ken C. L. Wong, Vandana Mukherjee, Convolutional autoencoder based model HistoCAE for segmentation of viable tumor regions in liver whole-slide images, Scientific Reports (11):139, 2021. (IF: 4.379)
Xiaoyuan Guo, Fusheng Wang, George Teodoro, Alton Farris, Jun Kong, Liver Steatosis Segmentation with Deep Learning Methods, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.24-27, Venice, Italy, April 2019.
|
-- In triple negative breast cancer (TNBC) treatment, early prediction of pathological complete response (PCR) from chemotherapy before surgical operations is crucial for optimal treatment planning. We propose a novel deep learning-based system to predict PCR to neoadjuvant chemotherapy for TNBC patients with multi-stained histopathology images of serial tissue sections. By first performing tumor cell detection and recognition in a cell detection module, we produce a set of feature maps that capture cell type, shape, and location information. Next, a newly designed spatial attention module integrates such feature maps with original pathology images in multiple stains for enhanced PCR prediction in a dedicated prediction module. We compare it with baseline models that either use a single-stained slide or have no spatial attention module in place. Additionally, the heatmaps generated from the spatial attention module can help pathologists in targeting tissue regions important for disease assessment. Our system presents high efficiency and effectiveness and improves interpretability, making it highly promising for immediate clinical and translational impact. 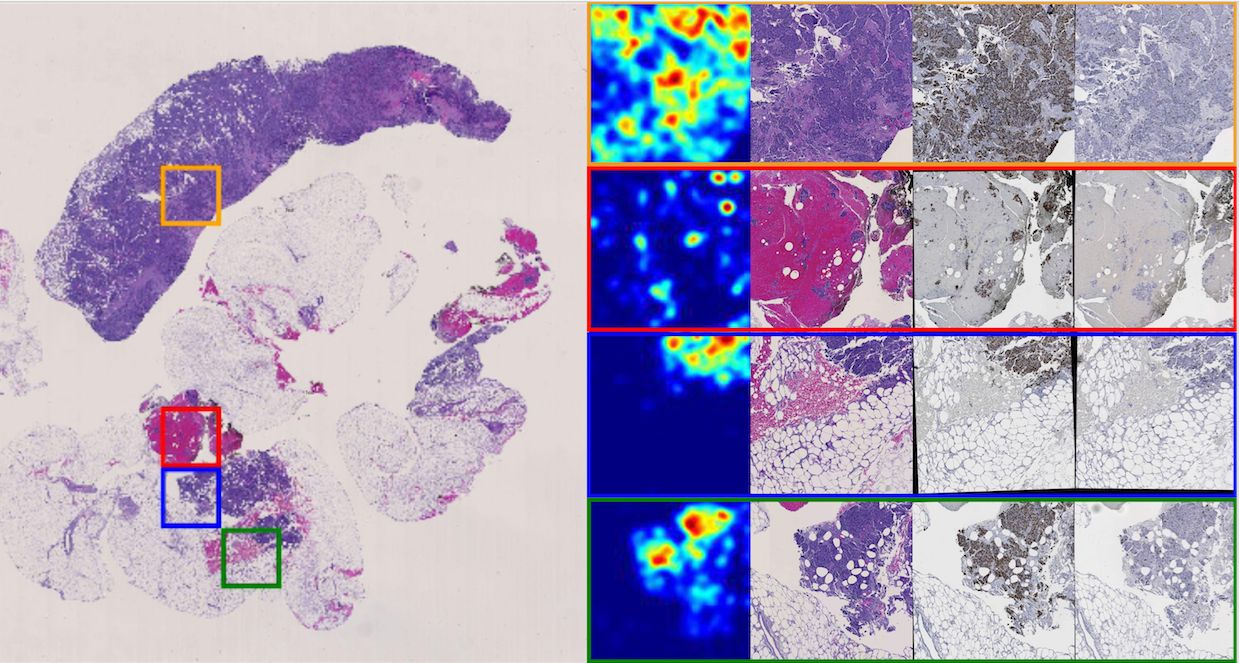
-- The pathological complete response to neoadjuvant chemotherapy is one good assessment measure in breast cancer treatment that is proven to be highly associated with patients' overall survival. While the accurate early prediction of PCR before neoadjuvant chemotherapy treatment is in high demand, the existing PCR prediction systems are too limited in accuracy to be deployed in real clinical settings. In this work, we proposed one convolutional neural network based system for predicting PCR from three different modalities of pathology images (hematoxylin and eosin (H&E), KI-67, and PHH3 stained) captured from adjacent tissue slices. One spatial attention mechanism is deployed to the system to guide it to pay attention to specific areas where pathologists consider important and informative. With integrated usage of different image modalities, the system can avoid the limitation of spatial homogeneity from original CNN. Our proposed model achieved a high patient-level accuracy, outperforming baselines and other state-of-the-art systems. Its inspiring performance suggests its great potential in real clinical usage. 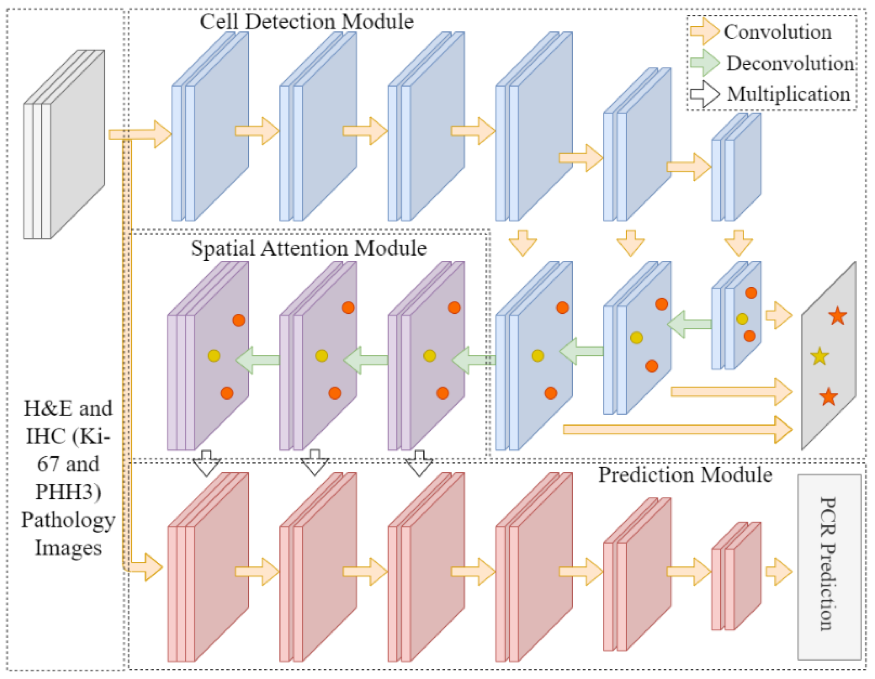
-- Oncotype DX Recurrence Score (RS) has been widely used to predict chemotherapy benefits in patients with estrogen receptor-positive breast cancer. Studies showed that the features used in Magee equations correlate with RS. We aimed to examine whether deep learning (DL)-based histology image analyses can enhance such correlations. We retrieved 382 cases with RS diagnosed between 2011 and 2015 from the Emory University and the Ohio State University. All patients received surgery. DL models were developed to detect nuclei of tumor cells and tumor-infiltrating lymphocytes (TILs) and segment tumor cell nuclei in hematoxylin and eosin (H&E) stained histopathology whole slide images (WSIs). Based on the DL-based analysis, we derived image features from WSIs, such as tumor cell number, TIL number variance, and nuclear grades. The entire patient cohorts were divided into one training set (125 cases) and two validation sets (82 and 175 cases) based on the data sources and WSI resolutions. The training set was used to train the linear regression models to predict RS. For prediction performance comparison, we used independent variables from Magee features alone or the combination of WSI-derived image and Magee features. Our results suggest that DL-based digital pathological features can enhance Magee feature correlation with RS. 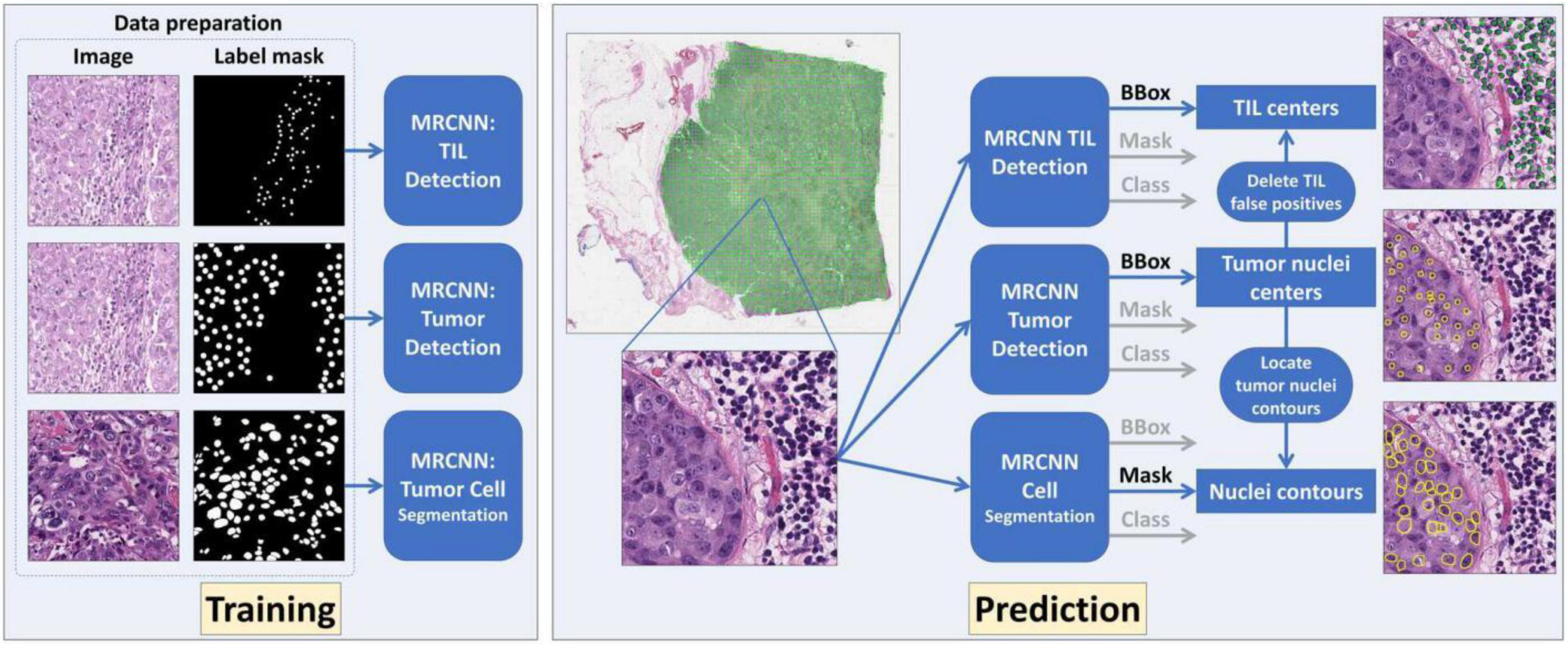
|
Hongyi Duanmu, Shristi Bhattarai, Hongxiao Li, Chia Cheng Cheng, Fusheng Wang, Georgia Teodoro, Emiel Janssen, Keerthi Gogineni, Preeti Subhedar, Ritu Aneja, Jun Kong, A Spatial Attention Guided Deep Learning System for Prediction of Pathological Complete Response Using Breast Cancer Histopathol- ogy Images, Bioinformatics, btac558, pp.1-8, 2022.
Hongyi Duanmu, Shristi Bhattarai, Hongxiao Li, Chia Cheng Cheng, Fusheng Wang, Georgia Teodoro, Emiel Janssen, Keerthi Gogineni, Preeti Subhedar, Ritu Aneja, Jun Kong, Spatial Attention-based Deep Learning System for Breast Cancer Pathological Complete Response Prediction with Serial Histopathology Images in Multiple Stains, International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), pp. 550-560, 2021.
Hongxiao Li, Jigang Wang, Zaibo Li, Melad Dababneh, Fusheng Wang, Peng Zhao, Geoffrey Smith, George Teodoro, Meijie Li, Jun Kong**, Xiaoxian Li** (senior author**), Deep learning-based pathology image analysis enhances Magee feature correlation with Oncotype DX Breast Recurrence Score, Frontiers in Medicine, Sec. Pathology, 2022.
Hanyi Yu, Fusheng Wang, Sung Bo Yoon, Robert Kauffman, Jens Wrammert, Adam Marcus, Jun Kong, Non-Gaussian Models for Object Motion Analysis with Time-lapse Fluorescence Microscopy Images, Modern Statistical Methods for Health Research. Accepted, Springer, In press, 2020.
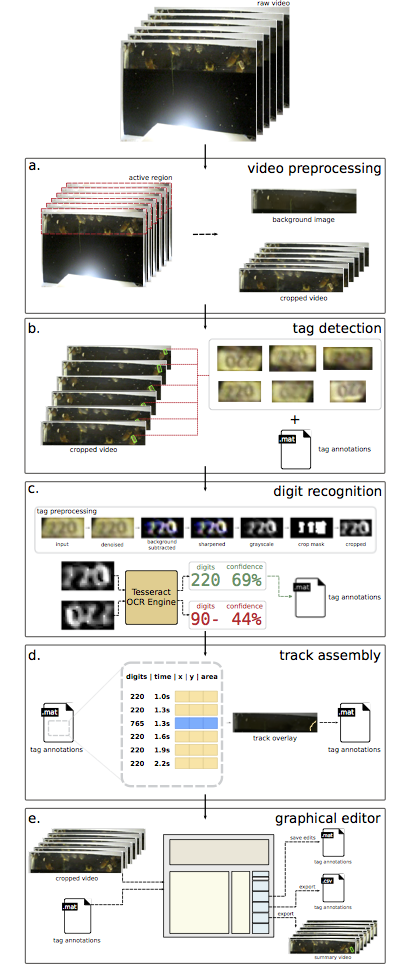
B.J. Rossetti, T. Dynes, B. Brosi, J.C. de Roode, Jun Kong, GRAPHITE: A Graphical Environment for Scalable Video Tracking of Animal Movement, Journal of Methods in Ecology and Evolution, vol:9 (4), pp. 956-964, 2018.
Blair Rossetti, Fusheng Wang, Pengyue Zhang, George Teodoro, Daniel Brat, Jun Kong, Dynamic Registration For Gigapixel Serial Whole Slide Images, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.424-428, Melbourne, Australia, April, 2017.
Yanhui Liang, Fusheng Wang, Pengyue Zhang, Joel Saltz, Daniel Brat, Jun Kong, Development of a Framework for Large Scale Three-Dimensional Pathology and Biomarker Imaging and Spatial Analytics, The American Medical Informatics Association (AMIA) Joint Summits on Translational Science, Accepted, 2017.
Jun Kong, P.Y. Zhang, Y.H. Liang, G. Teodoro, D.J. Brat, F.S. Wang, Robust Cell Segmentation for Histological Images of Glioblastoma, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp. 1041-1045, Prague, Czech Republic, April, 2016. (Oral).
P.Y. Zhang, Y.H. Liang, G. Teodoro, F.S. Wang, D.J. Brat, Jun Kong, Automated Level Set Segmentation of Histopathologic Objects with Sparse Shape Prior Support and Dynamic Occlusion Constraint, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.718-722, Melbourne, Australia, April 2017.
Jun Kong, F.S. Wang, G. Teodoro, Y.H. Liang, Y.Y. Zhu, C. Tucker-Burden, D.J. Brat, Automated Cell Recognition with 3D Fluorescence Microscopy Images, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.1212-1215, Brooklyn, NY, April 2015. (Oral)
P.Y. Zhang, F.S. Wang, G. Teodoro, Y.H. Liang, M. Roy, D.J. Brat, Jun Kong, Effective Cell Segmentation with Sparse Shape Prior and Dynamic Occlusion Constraint for Pathology Images, SPIE Journal of Medical Imaging, 6(1), 017502, pp.1-15, 2019.
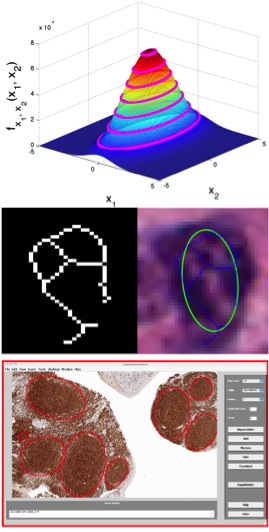
Jun Kong, K. Boyer, J. Saltz and K. Huang, A New Model-based Estimation of Ellipses for Object Representation, 31st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC2009), pp.3637-3640, Minneapolis, MN, September
2009.
Jun Kong, Michael Lee, Pelin Bagci, Puneet Sharma, Diego Martin, N. Volkan Adsay, Joel Saltz, and Brad Farris, Computer-based Image Analysis of Liver Steatosis with Large-scale Microscopy Imagery and Correlation with Magnetic Resonance Imaging Lipid Analysis, IEEE International Conference of bioinformatics and biomedicine (BIBM), pp. 333-338, Atlanta, GA, November, 2011. (Oral; Acceptance rate for regular paper: 19%)
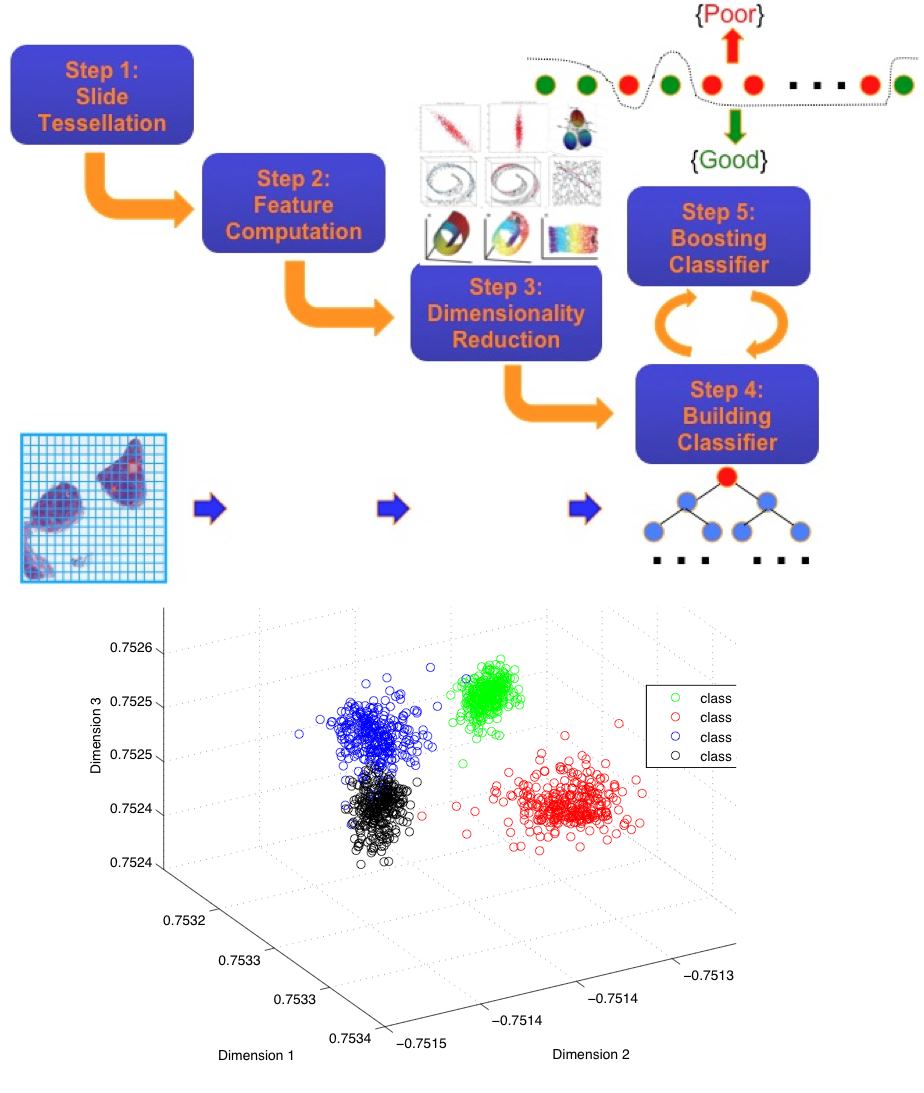
O. Sertel, Jun Kong, U.V. Catalyurek, G. Lozanski, J.H. Saltz and M. Gurcan, Histopathological Image Analysis for Follicular Lymphoma using Color Texture Models, Journal of Signal Processing Systems for Signal, Image, and Video Technology, DOI-10.1007/s11265-008-0201-y, Vol. 55, No. 1, pp.169-183, 2009.
O. Sertel, Jun Kong, G. Lozanski, U.V. Catalyurek, J.H. Saltz and M. Gurcan, Computerized microscopic image analysis of follicular lymphoma, Proc. SPIE Medical Imaging 2008, Vol. 6915, No. 1, 691535, San Diego, California, Feburary 2008. (Oral)
Jun Kong, Lee Cooper, Ashish Sharma, Tahsin Kurc, Daniel Brat, Joel Saltz, A new steerable pressure force for parametric deformable models, Proc. SPIE Medical Imaging 2011, Vol. 7962, 79623N, Orlando, Florida, February 2011.
Jun Kong, Lee Cooper, Tahsin Kurc, Daniel Brat, Joel Saltz, Towards Building Computerized Image Analysis Framework for Nucleus Discrimination in Microscopy Images of Diffuse Glioma, The 33rd International Conference of Engineering in Medicine and Biology Society(EMBC), pp. 6605-6608, Boston, MA, August, 2011. (Oral)
Jun Kong, Lee Cooper, Ashish Sharma, Tahsin Kurc, D. J. Brat and Joel H. Saltz, Texture Based Image Recognition in Microscopy Images of Diffuse Gliomas with Multi-class Gentle Boosting Mechanism, The 35th International Conference on Acoustics, Speech, and Signal
Processing (ICASSP), pp.457-460, Dallas, TX, March 2010. (Oral)
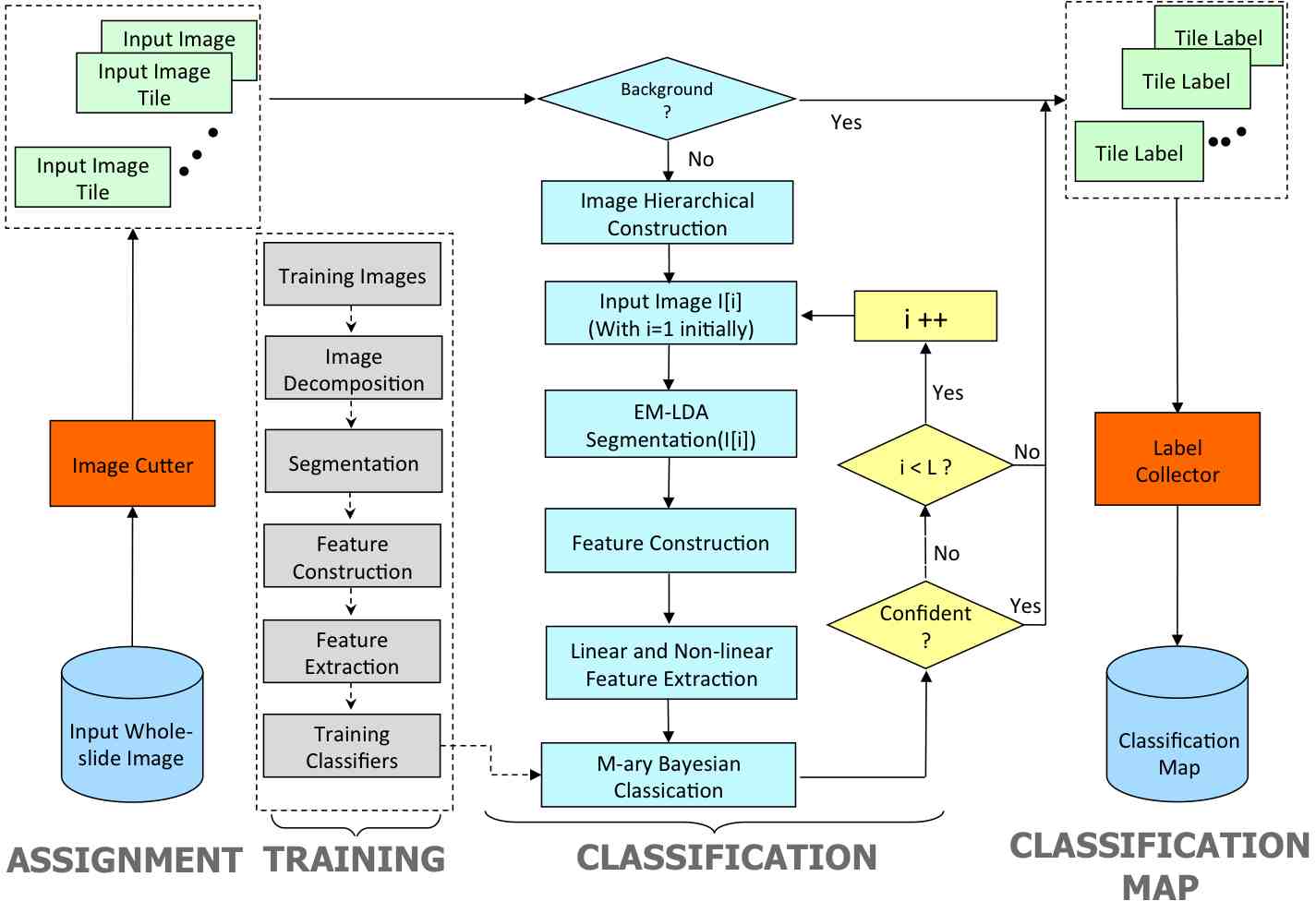
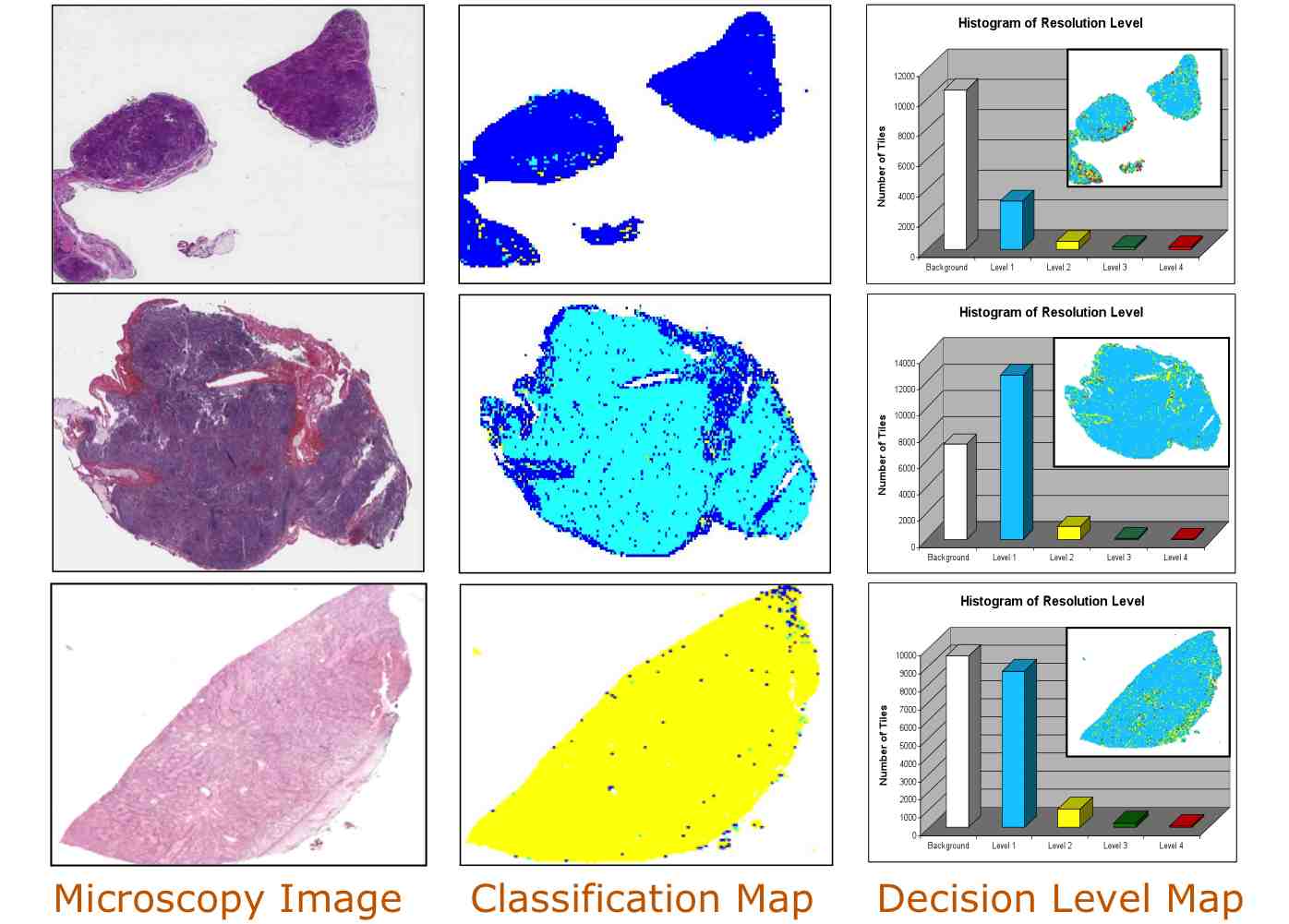
Jun Kong, O. Sertel, H. Shimada, K.L. Boyer, J.H. Saltz and M. Gurcan, Computeraided Evaluation of Neuroblastoma on Whole-slide Histology Images: Classifying Grade of Neuroblastic Differentiation, Journal of Pattern Recognition, Vol. 42, No. 6, pp.1080-1092, Jun 2009.
Jun Kong, H. Shimada, K.L. Boyer, J.H. Saltz and M. Gurcan, Image analysis for automated assessment of grade of neuroblastic differentiation, Proceedings of the Fourth IEEE
International Symposium on Biomedical Imaging (ISBI 2007), pp.61-64, Metro Washington DC, April 2007.
Mousumi Roy, Fusheng Wang, *Hoang Vo, Dejun Teng, George Teodoro, Alton B. Farris, Eduardo Castillo Lion, Miriam B. Vose, Jun Kong, Deep Learning Based Accurate Hepatic Steatosis Quantification for Histological Assessment of Liver Biopsies, Lab Investigation - Nature, 2020.(In press)
Mousumi Roy, Fusheng Wang, George Teodoro, Miriam Vos, Alton Brad Farris, Jun Kong, Segmentation of Overlapped Steatosis in Whole-Slide Liver Histopathology Microscopy Images, IEEE International Conference on Engineering in Medicine and Biology, pp.810-813, Honolulu, HI, 2018.
Xiaoyuan Guo, Fusheng Wang, George Teodoro, Alton Farris, Jun Kong, Liver Steatosis Segmentation with Deep Learning Methods, IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.24-27, Venice, Italy, April 2019.
Jun Kong, Michael Lee, Pelin Bagci, Puneet Sharma, Diego Martin, N. Volkan Adsay, Joel Saltz, and Brad Farris, Computer-based Image Analysis of Liver Steatosis with Large-scale Microscopy Imagery and Correlation with Magnetic Resonance Imaging Lipid Analysis, IEEE International Conference of bioinformatics and biomedicine (BIBM), pp. 333-338, Atlanta, GA, November, 2011. (Oral; Acceptance rate for regular paper: 19%)
Michael J. Lee, Pelin Bagci, Jun Kong, Miriam B. Vos, Puneet Sharma, Bobby Kalb, JoelSaltz, Diego R. Martin, N. Volkan Adsay, Alton B. Farris, Liver Steatosis Assessment: Correlations Among Pathology, Radiology, Clinical Data and Automated Image Analysis Software, Pathology-Research and
Practice, 209(6):pp.371-379, 2013.
M. Lee, P. Bagci, Jun Kong, M. Vos, V. Adsay, P. Sharma, D. Martin, A. Farris, Liver Steatosis Assessment: Correlations Between Pathology, Radiology, Clinical Data and Automated Image Analysis Software, The United States and Canadian Academy of Pathology’s 101st Annual Meeting, Vancouver, BC, Canada, March 2012.
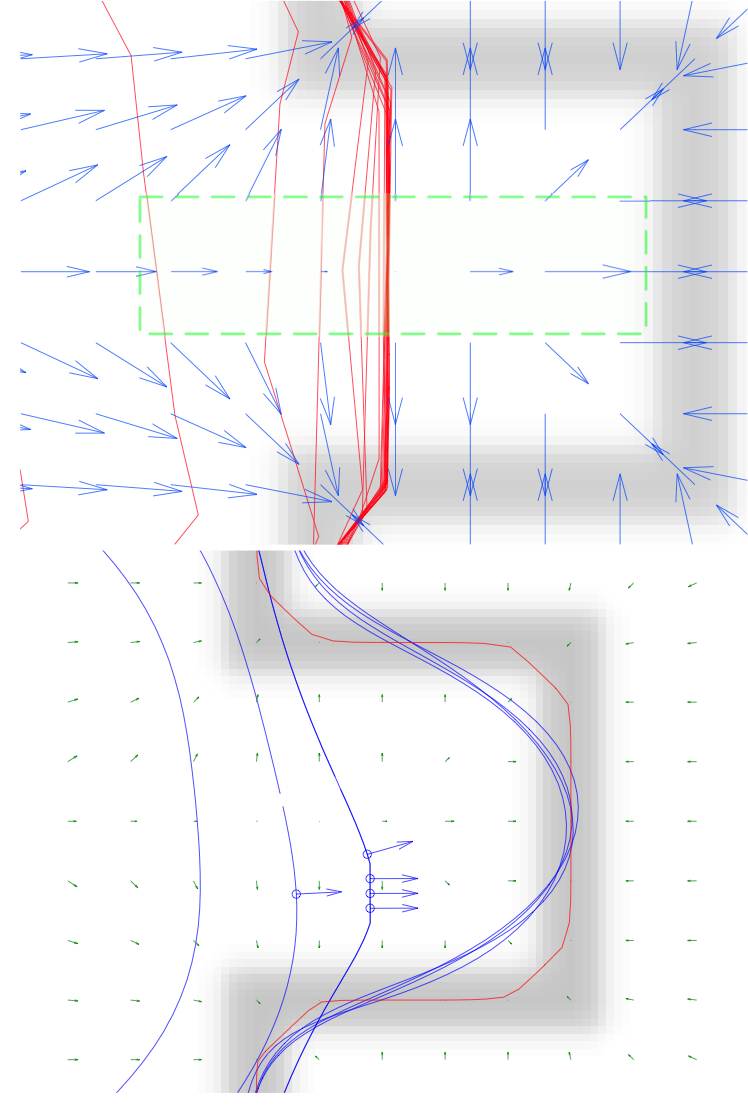
Y.H. Liang, F.S. Wang, D. Treanor, D. Magee, G. Teodoro, Y.Y. Zhu, Jun Kong, A 3D Primary Vessel Reconstruction Framework with Serial Microscopy Images, International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Part III, LNCS 9351, pp. 251-259, Munich, Germany, October, 2015.
Y.H. Liang, F.S. Wang, D. Treanor, D. Magee, G. Teodoro, Y.Y. Zhu, Jun Kong, Liver Whole Slide Image Analysis for 3D Vessel Reconstruction, International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp.182-185, Brooklyn, NY, April, 2015.
Y.H. Liang, F.S. Wang, D. Treanor, D. Magee, N. Roberts, G. Teodoro, Y.Y. Zhu, Jun Kong, A Framework for 3D Vessel Analysis using Whole Slide Images of Liver Tissue Sections, Accepted to International Journal of Computational Biology and Drug Design (IJCBDD) and International Conference on Intelligent Biology and Medicine (ICIBM), San Antonio, TX, 2014. (Travel Award)
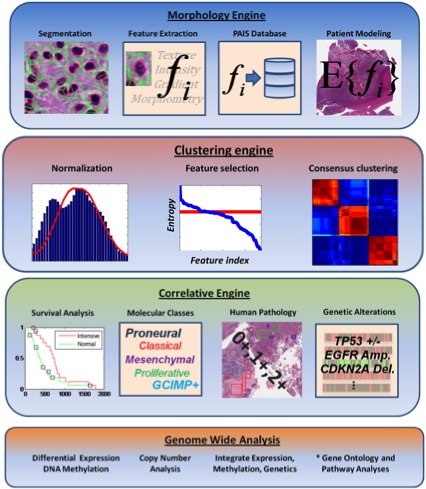
Jun Kong, Lee A.D. Cooper, Fusheng Wang, Jingjing Gao, George Teodoro, Lisa Scarpace, TomMikkelsen, Carlos S.Moreno, Joel H. Saltz, Daniel J. Brat, Generic, Computer-based
Morphometric Human Disease Classification Using Large Pathology Images Uncovers Signature
Molecular Correlates, PLoS One, 8(11), e81049. doi: 10.1371/journal.pone.0081049, November, 2013.
Jun Kong, Fusheng Wang, George Teodoro, Lee Cooper, Carlos Moreno, Tahsin Kurc, Tony Pan, Joel Saltz, and Daniel Brat, High-Performance Computational Analysis of Glioblastoma
Pathology Images with Database Support Identifies Molecular and Survival Correlates, IEEE International Conference of bioinformatics and biomedicine, pp.229-236, Shanghai, China, Decembe, 2013. (Oral; Acceptance rate for regular paper: 19.6%)
W. Caleb Rutledge, Jun Kong, Jingjing Gao, David Gutman, Lee Cooper, Christina Appin, Candace Chisolm, Yuna Park, Lisa Scarpace, Tom Mikkelsen, Mark Cohen, Ken Aldape,
Roger McLendon, Norman Lehman, Ryan Miller, Matthew J. Schniederjan, Cameron Brennan, Joel H. Saltz, Carlos S. Moreno, Daniel J. Brat, Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and enriched in the mesenchymal transcriptional class, Clinical Cancer Research, 19(18): pp.4951-4960, September, 2013.
Fusheng Wang, Jun Kong, Jingjing Gao, David Alder, Lee Cooper, Cristobal Vergara-Niedermayr, Zhengwen Zhou, Bryan Katigbak, Tahsin Kurc, Daniel Brat, Joel Saltz, A High Performance Spatial Database Based Approach for Pathology Imaging Algorithm Evaluation, Journal of Pathology Informatics, 4(1), 2013.
Lee Cooper, Jun Kong, David A. Gutman, Fusheng Wang, Doris Gao, Christina Appin, Sharath R. Cholleti, Tony C. Pan, Ashish Sharma, Lisa Scarpace, Tom Mikkelsen, Tahsin Kurc, Carlos S. Moreno, Daniel J. Brat, Joel H. Saltz, Integrated Morphologic Analysis for the Identification and Characterization of Disease Subtypes, Journal of American Medical Informatics Association, 19(2):317-323, 2012.
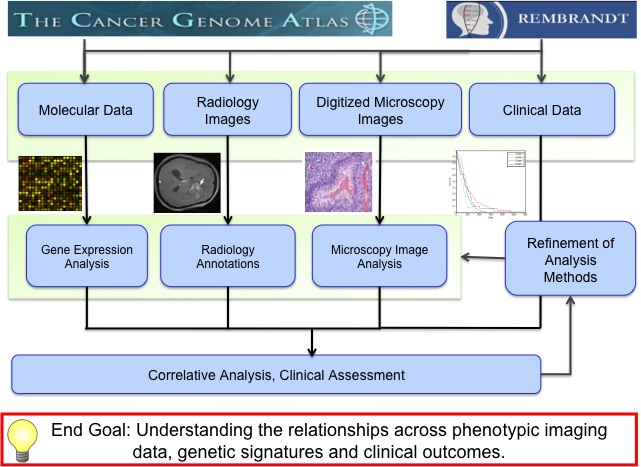
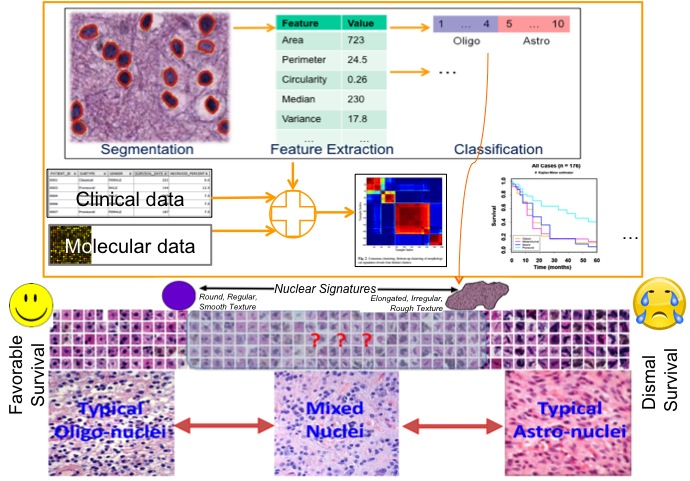
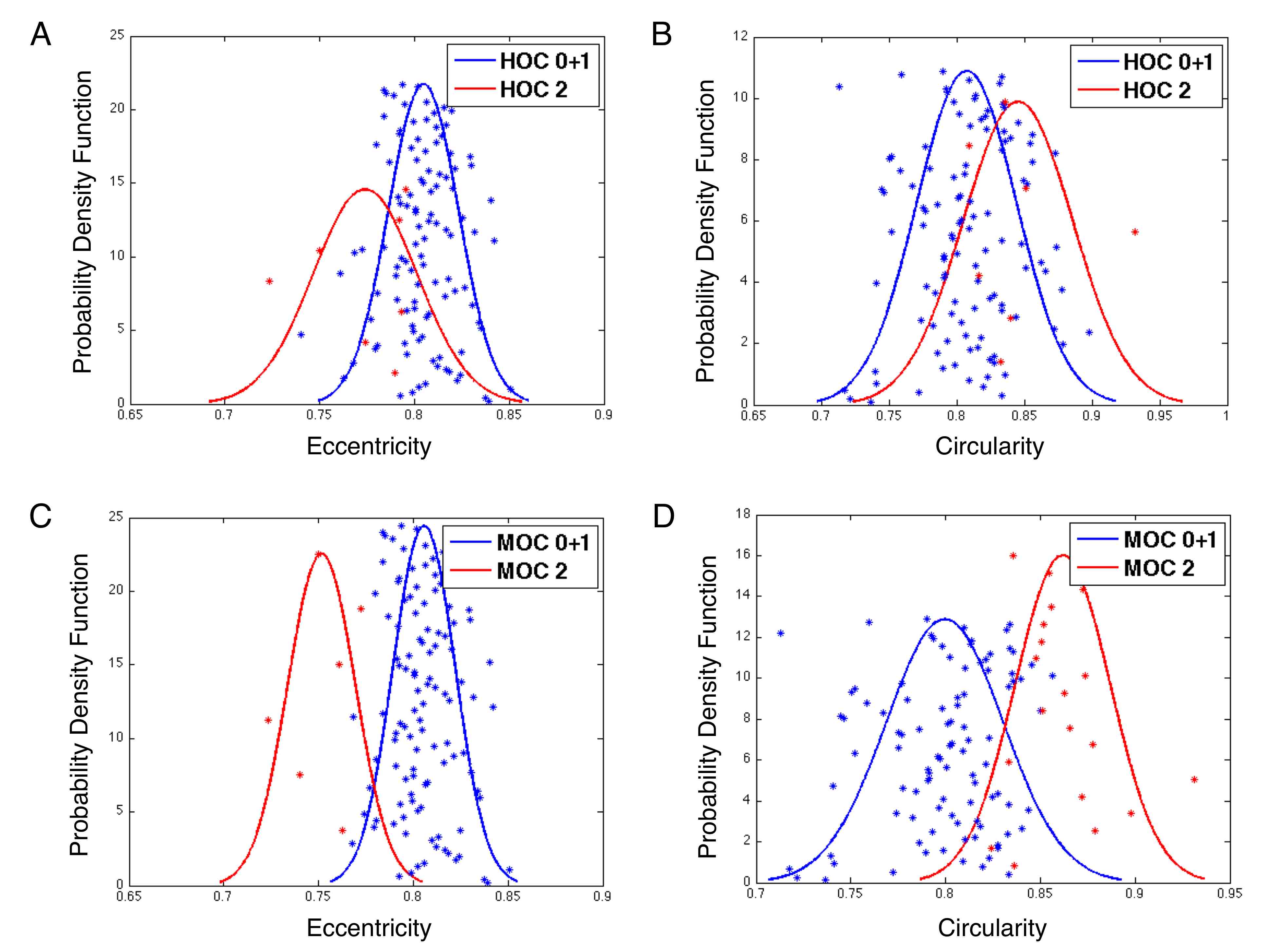
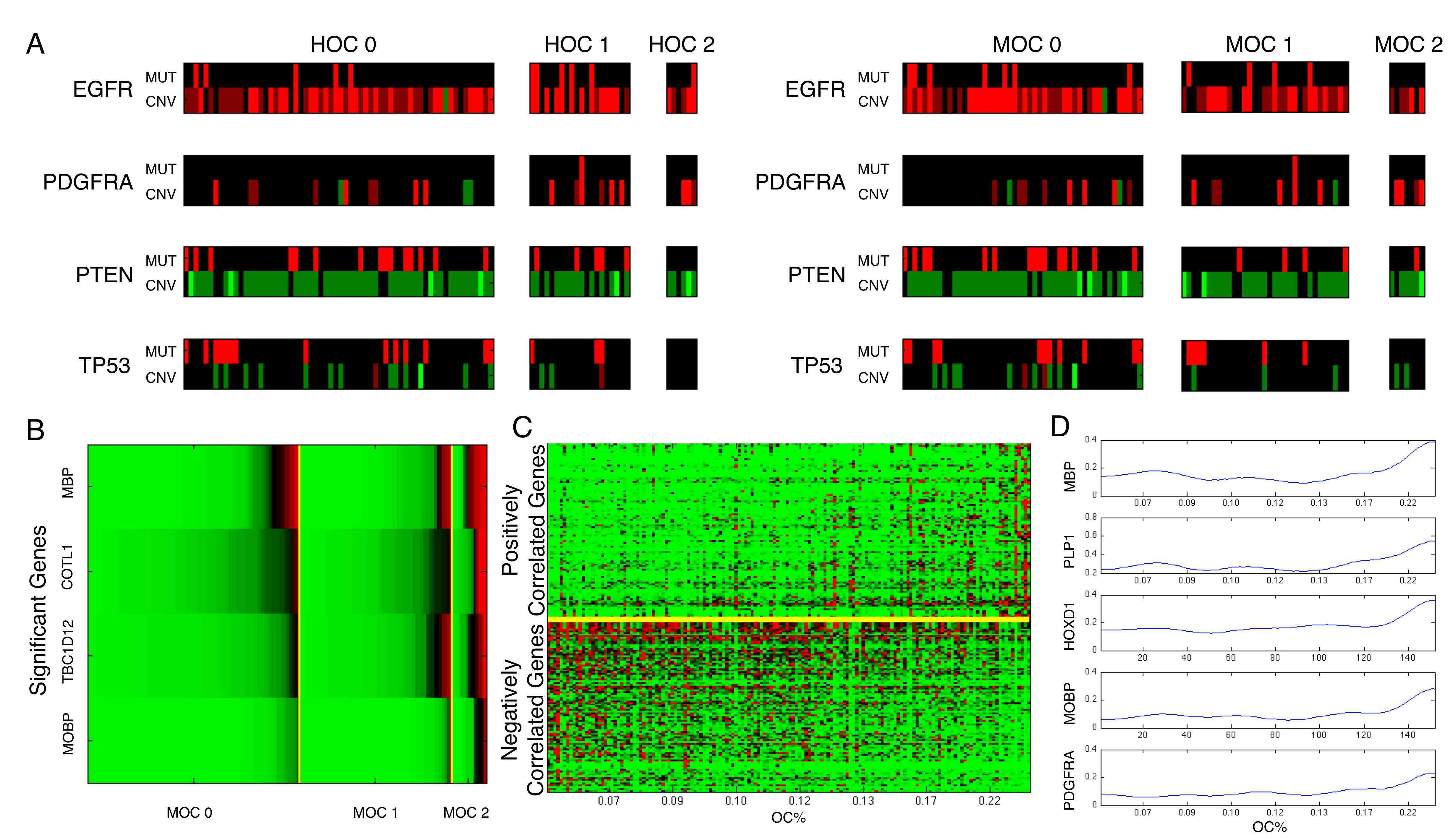
Jun Kong, Lee A.D. Cooper, Fusheng Wang, David A. Gutman, Jingjing Gao, Candace Chisolm, Ashish Sharma, Tony C. Pan, Erwin G. Van Meir, Tahsin M. Kurc, Carlos S. Moreno, Joel H. Saltz and Daniel J. Brat, Integrative, Multi-modal Analysis of Glioblastoma Using
TCGA Molecular Data, Pathology Images and Clinical Outcomes, IEEE Transactions on Biomedical Engineering, Vol. 58, No. 12, pp. 3469-3474, Dec 2011. (article on journal cover page)
Jun Kong, Lee Cooper, Carlos Moreno, Fusheng Wang, Tahsin Kurc, Joel Saltz, Daniel Brat, In Silico Analysis of Nuclei in Glioblastoma using Large-scale Microscopy Images Improves
Prediction of Treatment Response, The 33rd International Conference of Engineering in Medicine and Biology Society(EMBC), pp. 87-90, Boston, MA, August, 2011.
Jun Kong, Lee Cooper, Fusheng Wang, Candace Chisolm, Carlos Moreno, Tahsin Kurc, Patrick Widener, Daniel Brat, Joel Saltz, A Comprehensive Framework for Classification of Nuclei in Digital Microscopy Imaging: An Application to Diffuse Gliomas, The 8th International Symposium on Biomedical Imaging(ISBI), pp. 2128-2131, Chicago, Illinois, March, 2011. (Oral)
Lee Cooper, Jun Kong, Fusheng Wang, Tahsin Kurc, Carlos Moreno, Daniel Brat, Joel Saltz, Morphological Signatures and Genomic Correlates in Glioblastoma, The 8th International Symposium on Biomedical Imaging(ISBI), pp. 1624-1627, Chicago, Illinois, March, 2011. (Oral)
Lee Cooper, Jun Kong, David Gutman, Fusheng Wang, Sharath Cholleti, Tony Pan, Patrick Widener, Ashish Sharma, Tom Mikkelsen, Adam Flanders, Daniel Rubin, Erwin Van Meir, Tahsin Kurc, Carlos Moreno, Daniel Brat, Joel Saltz, An Integrative Approach for In Silico Glioma Research , IEEE Transactions on Biomedical Engineering, Vol. 57, No. 10, pp. 2617-2621, October 2010.
Pathological Image Segmentation for Neuroblastoma Using the GPU, Proceedings of the Fifth IEEE International Symposium on Biomedical Imaging (ISBI 2008), pp.296-299, Paris, France, May
2008. (Oral)
Computeraided Evaluation of Neuroblastoma on Whole-slide Histology Images: Classifying Grade of Neuroblastic Differentiation, Journal of Pattern Recognition, Vol. 42, No. 6, pp.1080-1092, Jun 2009.
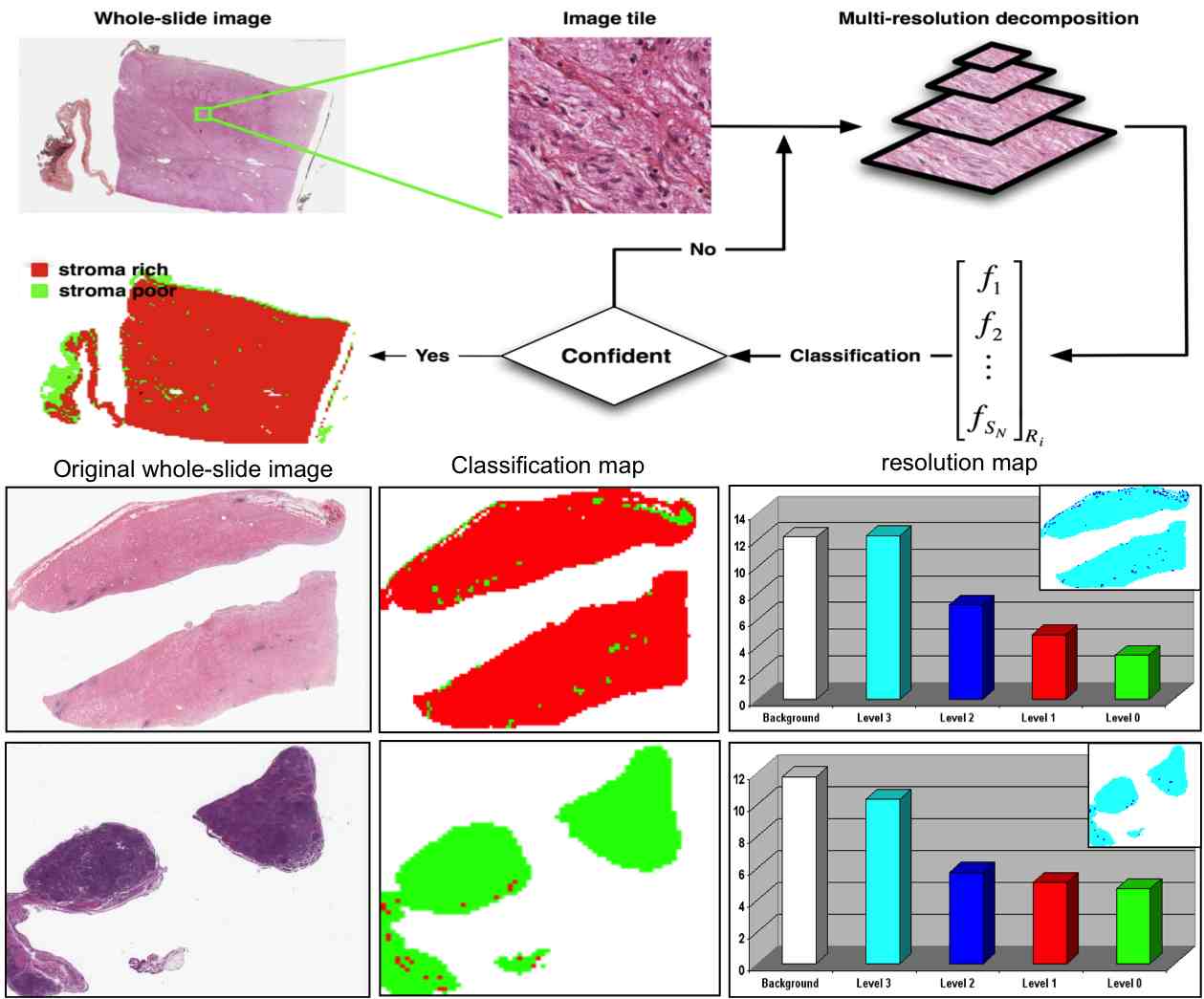
Computeraided Prognosis of Neuroblastoma on Whole-slide Images: Classification of Stromal Development, Journal of Pattern Recognition, Vol. 42, No. 6, pp.1093-1103, Jun 2009.
Computer assisted Grading of Neuroblastic Differentiation, Journal of Archive Pathology and Laboratory Medicine, Vol. 132, No. 6, pp. 903-904, June, 2008.
A multiresolution image analysis system for computer-assisted grading of neuroblastoma differentiation, Proc. SPIE Medical Imaging 2008, Vol. 6915, No. 1, 69151T, San Diego, California, Feburary 2008.
Computer-aided prognosis of neuroblastoma: Classification of stromal development on whole-slide images,
Proc. SPIE Medical Imaging 2008, Vol. 6915, No. 1, 69150P, San Diego, California, Feburary 2008.
Computerized Pathological Image Analysis for Neuroblastoma Prognosis, Proceedings of the Annual Symposium of American Medical Informatics Association 2007 (AMIA 2007), pp.304-308, Chicago, IL, November 2007.
Efficient Processing of Pathological Images Using the Grid: Computer-Aided Prognosis of Neuroblastoma, Proceedings of the Fifth IEEE International Conference on Challenges of Large Applications in Distributed Environments (CLADE 2007), pp.35-41, Monterey Bay, CA, June 2007.
Computeraided Grading of Neuroblastic Differentiation: Multi-resolution and Multi-classifier Approach,
Proceedings of the IEEE International Conference on Image Processing(ICIP 2007), pp.525-528, San Antonio, TX, September 2007.
Image analysis for automated assessment of grade of neuroblastic differentiation, Proceedings of the Fourth IEEE
International Symposium on Biomedical Imaging (ISBI 2007), pp.61-64, Metro Washington DC, April 2007.